Positively regulated bacterial expression systems
- PMID: 21261879
- PMCID: PMC3815419
- DOI: 10.1111/j.1751-7915.2008.00048.x
Positively regulated bacterial expression systems
Abstract
Regulated promoters are useful tools for many aspects related to recombinant gene expression in bacteria, including for high-level expression of heterologous proteins and for expression at physiological levels in metabolic engineering applications. In general, it is common to express the genes of interest from an inducible promoter controlled either by a positive regulator or by a repressor protein. In this review, we discuss established and potentially useful positively regulated bacterial promoter systems, with a particular emphasis on those that are controlled by the AraC-XylS family of transcriptional activators. The systems function in a wide range of microorganisms, including enterobacteria, soil bacteria, lactic bacteria and streptomycetes. The available systems that have been applied to express heterologous genes are regulated either by sugars (L-arabinose, L-rhamnose, xylose and sucrose), substituted benzenes, cyclohexanone-related compounds, ε-caprolactam, propionate, thiostrepton, alkanes or peptides. It is of applied interest that some of the inducers require the presence of transport systems, some are more prone than others to become metabolized by the host and some have been applied mainly in one or a limited number of species. Based on bioinformatics analyses, the AraC-XylS family of regulators contains a large number of different members (currently over 300), but only a small fraction of these, the XylS/Pm, AraC/P(BAD), RhaR-RhaS/rhaBAD, NitR/PnitA and ChnR/Pb regulator/promoter systems, have so far been explored for biotechnological applications.
© 2008 The Authors; Journal compilation © 2008 Society for Applied Microbiology and Blackwell Publishing Ltd.
Figures


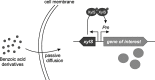
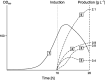
References
-
- Adhya S., Garges S. Positive control. J Biol Chem. 1990;265:10797–10800. - PubMed
-
- Ali N., Herron P.R., Evans M.C., Dyson P.J. Osmotic regulation of the Streptomyces lividans thiostrepton‐inducible promoter PtipA. Microbiology. 2002;148:381–390. - PubMed
-
- Altenbuchner J., Mattes R. Escherichia coli. In: Gellissen G., editor. WILEY‐VCH Verlag GmbH and Co. KGaA; 2005. pp. 7–43.
-
- Axelsson L., Lindstad G., Naterstad K. Development of an inducible gene expression system for Lactobacillus sakei. Lett Appl Microbiol. 2003;37:1–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

