Interplay of metagenomics and in vitro compartmentalization
- PMID: 21261880
- PMCID: PMC3815420
- DOI: 10.1111/j.1751-7915.2008.00057.x
Interplay of metagenomics and in vitro compartmentalization
Abstract
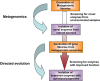
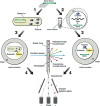
In recent years, the application of approaches for harvesting DNA from the environment, the so-called, 'metagenomic approaches' has proven to be highly successful for the identification, isolation and generation of novel enzymes. Functional screening for the desired catalytic activity is one of the key steps in mining metagenomic libraries, as it does not rely on sequence homology. In this mini-review, we survey high-throughput screening tools, originally developed for directed evolution experiments, which can be readily adapted for the screening of large libraries. In particular, we focus on the use of in vitro compartmentalization (IVC) approaches to address potential advantages and problems the merger of culture-independent and IVC techniques might bring on the mining of enzyme activities in microbial communities.
© 2008 The Authors; Journal compilation © 2008 Society for Applied Microbiology and Blackwell Publishing Ltd.
Figures

References
-
- Aharoni A., Amitai G., Bernath K., Magdassi S., Tawfik D.S. High‐throughput screening of enzyme libraries: thiolactonases evolved by fluorescence‐activated sorting of single cells in emulsion compartments. Chem Biol. 2005a;12:1281–1289. - PubMed
-
- Aharoni A., Griffiths A.D., Tawfik D.S. High‐throughput screens and selections of enzyme encoding genes. Curr Opin Chem Biol. 2005b;9:210–216. - PubMed
-
- Andexer J., Guterl J.K., Pohl M., Eggert T. A high‐throughput screening assay for hydroxynitrile lyase activity. Chem Commun (Camb) 2006;40:4201–4203. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

