Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain
- PMID: 21262798
- PMCID: PMC3038701
- DOI: 10.1073/pnas.1018921108
Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain
Abstract
The plasma membrane sodium/proton exchanger Salt-Overly-Sensitive 1 (SOS1) is a critical salt tolerance determinant in plants. The SOS2-SOS3 calcium-dependent protein kinase complex up-regulates SOS1 activity, but the mechanistic details of this crucial event remain unresolved. Here we show that SOS1 is maintained in a resting state by a C-terminal auto-inhibitory domain that is the target of SOS2-SOS3. The auto-inhibitory domain interacts intramolecularly with an adjacent domain of SOS1 that is essential for activity. SOS1 is relieved from auto-inhibition upon phosphorylation of the auto-inhibitory domain by SOS2-SOS3. Mutation of the SOS2 phosphorylation and recognition site impeded the activation of SOS1 in vivo and in vitro. Additional amino acid residues critically important for SOS1 activity and regulation were identified in a genetic screen for hypermorphic alleles.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
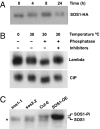

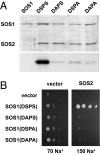
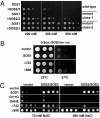
References
-
- Abrol IP, Yadav JSP, Massoud FI. FAO Soils Bull. Food and Agriculture Organization (Rome); 1988. Salt-affected soils and their management.
-
- Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

