The cell-adhesion G protein-coupled receptor BAI3 is a high-affinity receptor for C1q-like proteins
- PMID: 21262840
- PMCID: PMC3038708
- DOI: 10.1073/pnas.1019577108
The cell-adhesion G protein-coupled receptor BAI3 is a high-affinity receptor for C1q-like proteins
Abstract
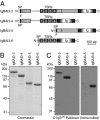
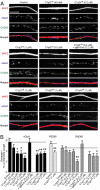
C1q-like genes (C1ql1-C1ql4) encode small, secreted proteins that are expressed in differential patterns in the brain but whose receptors and functions remain unknown. BAI3 protein, in contrast, is a member of the cell-adhesion class of G protein-coupled receptors that are expressed at high levels in the brain but whose ligands have thus far escaped identification. Using a biochemical approach, we show that all four C1ql proteins bind to the extracellular thrombospondin-repeat domain of BAI3 with high affinity, and that this binding is mediated by the globular C1q domains of the C1ql proteins. Moreover, we demonstrate that addition of submicromolar concentrations of C1ql proteins to cultured neurons causes a significant decrease in synapse density, and that this decrease was prevented by simultaneous addition of the thrombospondin-repeat fragment of BAI3, which binds to C1ql proteins. Our data suggest that C1ql proteins are secreted signaling molecules that bind to BAI3 and act, at least in part, to regulate synapse formation and/or maintenance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Yona S, Lin HH, Siu WO, Gordon S, Stacey M. Adhesion-GPCRs: Emerging roles for novel receptors. Trends Biochem Sci. 2008;33:491–500. - PubMed
-
- Lagerström MC, Schiöth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–357. - PubMed
-
- Bjarnadóttir TK, et al. The human and mouse repertoire of the adhesion family of G protein-coupled receptors. Genomics. 2004;84:23–33. - PubMed
-
- Krasnoperov V, et al. Post-translational proteolytic processing of the calcium-independent receptor of alpha-latrotoxin (CIRL), a natural chimera of the cell adhesion protein and the G protein-coupled receptor: Role of the G protein-coupled receptor proteolysis site (GPS) motif. J Biol Chem. 2002;277:46518–46526. - PubMed
-
- Lin HH, et al. Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. J Biol Chem. 2004;279:31823–31832. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

