The double face of the histone variant H3.3
- PMID: 21263457
- PMCID: PMC3193428
- DOI: 10.1038/cr.2011.14
The double face of the histone variant H3.3
Abstract
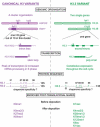
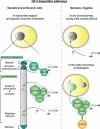
Histone proteins wrap DNA to form nucleosome particles that compact eukaryotic genomes while still allowing access for cellular processes such as transcription, replication and DNA repair. Histones exist as different variants that have evolved crucial roles in specialized functions in addition to their fundamental role in packaging DNA. H3.3--a conserved histone variant that is structurally very close to the canonical histone H3--has been associated with active transcription. Furthermore, its role in histone replacement at active genes and promoters is highly conserved and has been proposed to participate in the epigenetic transmission of active chromatin states. Unexpectedly, recent data have revealed accumulation of this specific variant at silent loci in pericentric heterochromatin and telomeres, raising questions concerning the actual function of H3.3. In this review, we describe the known properties of H3.3 and the current view concerning its incorporation modes involving particular histone chaperones. Finally, we discuss the functional significance of the use of this H3 variant, in particular during germline formation and early development in different species.
Figures


References
-
- Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. - PubMed
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. - PubMed
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
-
- Zweidler A.Core histone variants of the mouse: primary structure and differential expressionIn: JL Stein GS Stein, WF Marzluff, eds. Vol. Chapter 14, Histones Genes, Structure, Organization and Regulation. John Wiley & sons, 1984339–371.
-
- Talbert PB, Henikoff S. Histone variants - ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11:264–275. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

