The ESCRT system is required for hepatitis C virus production
- PMID: 21264300
- PMCID: PMC3019154
- DOI: 10.1371/journal.pone.0014517
The ESCRT system is required for hepatitis C virus production
Abstract
Background: Recently, lipid droplets have been found to be involved in an important cytoplasmic organelle for hepatitis C virus (HCV) production. However, the mechanisms of HCV assembly, budding, and release remain poorly understood. Retroviruses and some other enveloped viruses require an endosomal sorting complex required for transport (ESCRT) components and their associated proteins for their budding process.
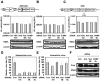
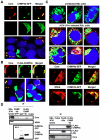
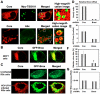
Methodology/principal findings: To determine whether or not the ESCRT system is needed for HCV production, we examined the infectivity of HCV or the Core levels in culture supernatants as well as HCV RNA levels in HuH-7-derived RSc cells, in which HCV-JFH1 can infect and efficiently replicate, expressing short hairpin RNA or siRNA targeted to tumor susceptibility gene 101 (TSG101), apoptosis-linked gene 2 interacting protein X (Alix), Vps4B, charged multivesicular body protein 4b (CHMP4b), or Brox, all of which are components of the ESCRT system. We found that the infectivity of HCV in the supernatants was significantly suppressed in these knockdown cells. Consequently, the release of the HCV Core into the culture supernatants was significantly suppressed in these knockdown cells after HCV-JFH1 infection, while the intracellular infectivity and the RNA replication of HCV-JFH1 were not significantly affected. Furthermore, the HCV Core mostly colocalized with CHMP4b, a component of ESCRT-III. In this context, HCV Core could bind to CHMP4b. Nevertheless, we failed to find the conserved viral late domain motif, which is required for interaction with the ESCRT component, in the HCV-JFH1 Core, suggesting that HCV Core has a novel motif required for HCV production.
Conclusions/significance: These results suggest that the ESCRT system is required for infectious HCV production.
Conflict of interest statement
Figures


References
-
- Boulant S, Douglas MW, Moody L, Budkowska A, Targett-Adams P, et al. Hepatitis C virus core protein induces lipid droplet redistribution in a microtubule- and dynein-dependent manner. Traffic. 2008;9:1268–1282. - PubMed
-
- Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, et al. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–1097. - PubMed
-
- Shi ST, Polyak SJ, Tu H, Taylor DR, Gretch DR, et al. Hepatitis C virus NS5A colocalizes with the core protein on lipid droplets and interacts with apolipoproteins. Virology. 2002;292:198–210. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

