Structure-based analysis of five novel disease-causing mutations in 21-hydroxylase-deficient patients
- PMID: 21264314
- PMCID: PMC3019215
- DOI: 10.1371/journal.pone.0015899
Structure-based analysis of five novel disease-causing mutations in 21-hydroxylase-deficient patients
Abstract
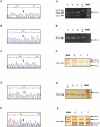
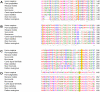
Congenital adrenal hyperplasia (CAH) due to 21-hydroxylase deficiency is the most frequent inborn error of metabolism, and accounts for 90-95% of CAH cases. The affected enzyme, P450C21, is encoded by the CYP21A2 gene, located together with a 98% nucleotide sequence identity CYP21A1P pseudogene, on chromosome 6p21.3. Even though most patients carry CYP21A1P-derived mutations, an increasing number of novel and rare mutations in disease causing alleles were found in the last years. In the present work, we describe five CYP21A2 novel mutations, p.R132C, p.149C, p.M283V, p.E431K and a frameshift g.2511_2512delGG, in four non-classical and one salt wasting patients from Argentina. All novel point mutations are located in CYP21 protein residues that are conserved throughout mammalian species, and none of them were found in control individuals. The putative pathogenic mechanisms of the novel variants were analyzed in silico. A three-dimensional CYP21 structure was generated by homology modeling and the protein design algorithm FoldX was used to calculate changes in stability of CYP21A2 protein. Our analysis revealed changes in protein stability or in the surface charge of the mutant enzymes, which could be related to the clinical manifestation found in patients.
Conflict of interest statement
Figures
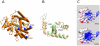
Similar articles
-
Functional studies of p.R132C, p.R149C, p.M283V, p.E431K, and a novel c.652-2A>G mutations of the CYP21A2 gene.PLoS One. 2014 Mar 25;9(3):e92181. doi: 10.1371/journal.pone.0092181. eCollection 2014. PLoS One. 2014. PMID: 24667412 Free PMC article.
-
Three novel CYP21A2 mutations and their protein modelling in patients with classical 21-hydroxylase deficiency from northeastern Iran.Clin Endocrinol (Oxf). 2007 Sep;67(3):335-41. doi: 10.1111/j.1365-2265.2007.02886.x. Epub 2007 Jun 15. Clin Endocrinol (Oxf). 2007. PMID: 17573904
-
A rational, non-radioactive strategy for the molecular diagnosis of congenital adrenal hyperplasia due to 21-hydroxylase deficiency.Gene. 2013 Sep 10;526(2):239-45. doi: 10.1016/j.gene.2013.03.082. Epub 2013 Apr 6. Gene. 2013. PMID: 23570880
-
CYP21A2 intronic variants causing 21-hydroxylase deficiency.Metabolism. 2017 Jun;71:46-51. doi: 10.1016/j.metabol.2017.03.003. Epub 2017 Mar 9. Metabolism. 2017. PMID: 28521877 Review.
-
Molecular diagnosis of congenital adrenal hyperplasia due to 21-hydroxylase deficiency: an update of new CYP21A2 mutations.Clin Chem Lab Med. 2010 Aug;48(8):1057-62. doi: 10.1515/CCLM.2010.239. Clin Chem Lab Med. 2010. PMID: 20482300 Review.
Cited by
-
Functional studies of p.R132C, p.R149C, p.M283V, p.E431K, and a novel c.652-2A>G mutations of the CYP21A2 gene.PLoS One. 2014 Mar 25;9(3):e92181. doi: 10.1371/journal.pone.0092181. eCollection 2014. PLoS One. 2014. PMID: 24667412 Free PMC article.
-
Congenital Adrenal Hyperplasia (CAH) due to 21-Hydroxylase Deficiency: A Comprehensive Focus on 233 Pathogenic Variants of CYP21A2 Gene.Mol Diagn Ther. 2018 Jun;22(3):261-280. doi: 10.1007/s40291-018-0319-y. Mol Diagn Ther. 2018. PMID: 29450859 Review.
-
Clinical outcomes and characteristics of P30L mutations in congenital adrenal hyperplasia due to 21-hydroxylase deficiency.Endocrine. 2020 Aug;69(2):262-277. doi: 10.1007/s12020-020-02323-3. Epub 2020 May 5. Endocrine. 2020. PMID: 32367336 Free PMC article. Review.
-
Structure-based activity prediction of CYP21A2 stability variants: A survey of available gene variations.Sci Rep. 2016 Dec 14;6:39082. doi: 10.1038/srep39082. Sci Rep. 2016. PMID: 27966633 Free PMC article.
-
Enhancing human spermine synthase activity by engineered mutations.PLoS Comput Biol. 2013;9(2):e1002924. doi: 10.1371/journal.pcbi.1002924. Epub 2013 Feb 28. PLoS Comput Biol. 2013. PMID: 23468611 Free PMC article.
References
-
- New MI, White PC, Pang S, Dupont B, Speiser PW. The adrenal hyperplasias. Scriver CR, Beaudet AL, Sly S, Valle D, editors. 1989:1881–1917. The Metabolic Basis of Inherited Disease, McGraw-Hill, New York. 6th edn,
-
- Miller WL. Genetics, diagnosis and management of 21-hydroxylase deficiency. J Clin Endocrinol Metab. 1994;78:241–246. - PubMed
-
- Pang S, Shook MK. Current status of neonatal screening for congenital adrenal hyperplasia. Curr Opin Pediatr. 1997;9:419–23. - PubMed
-
- Therell BL. Newborn screening for congenital adrenal hyperplasia. Endocrinol Metab Clin North Am. 2001;30:15–30. - PubMed
-
- Van der Kamp HJ, Wit JM. Neonatal screening for congenital adrenal hyperplasia. Eur J Endocrinol. 2004;151:U71–U75. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

