Detection of a single identical cytomegalovirus (CMV) strain in recently seroconverted young women
- PMID: 21264339
- PMCID: PMC3018470
- DOI: 10.1371/journal.pone.0015949
Detection of a single identical cytomegalovirus (CMV) strain in recently seroconverted young women
Abstract
Background: Infection with multiple CMV strains is common in immunocompromised hosts, but its occurrence in normal hosts has not been well-studied.
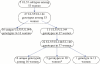
Methods: We analyzed CMV strains longitudinally in women who acquired CMV while enrolled in a CMV glycoprotein B (gB) vaccine trial. Sequencing of four variable genes was performed in samples collected from seroconversion and up to 34 months thereafter.
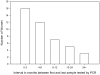
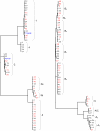
Results: 199 cultured isolates from 53 women and 65 original fluids from a subset of 19 women were sequenced. 51 women were infected with one strain each without evidence for genetic drift; only two women shed multiple strains. Genetic variability among strains increased with the number of sequenced genetic loci. Nevertheless, 13 of 53 women proved to be infected with an identical CMV strain based on sequencing at all four variable genes. CMV vaccine did not alter the degree of genetic diversity amongst strains.
Conclusions: Primary CMV infection in healthy women nearly always involves shedding of one strain that remains stable over time. Immunization with CMVgB-1 vaccine strain is not selective against specific strains. Although 75% of women harbored their unique strain, or a strain shared with only one other woman, 25% shared a single common strain, suggesting that this predominant strain with a particular combination of genetic loci is advantageous in this large urban area.
Conflict of interest statement
Figures
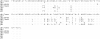


References
-
- Bale JF, Jr, Petheram SJ, Souza IE, Murph JR. Cytomegalovirus reinfection in young children. J Pediatr. 1996;128:347–352. - PubMed
-
- Puchhammer-Stockl E, Gorzer I, Zoufaly A, Jaksch P, Bauer CC, et al. Emergence of multiple cytomegalovirus strains in blood and lung of lung transplant recipients. Transplantation. 2006;81:187–194. - PubMed
-
- Arav-Boger R, Willoughby RE, Pass RF, Zong JC, Jang WJ, et al. Polymorphisms of the cytomegalovirus (CMV)-encoded tumor necrosis factor-alpha and beta-chemokine receptors in congenital CMV disease. J Infect Dis. 2002;186:1057–1064. - PubMed
-
- Trincado DE, Scott GM, White PA, Hunt C, Rasmussen L, et al. Human cytomegalovirus strains associated with congenital and perinatal infections. J Med Virol. 2000;61:481–487. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

