Evidence of activity-specific, radial organization of mitotic chromosomes in Drosophila
- PMID: 21264350
- PMCID: PMC3019107
- DOI: 10.1371/journal.pbio.1000574
Evidence of activity-specific, radial organization of mitotic chromosomes in Drosophila
Abstract
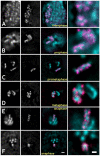
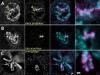
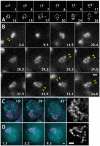
The organization and the mechanisms of condensation of mitotic chromosomes remain unsolved despite many decades of efforts. The lack of resolution, tight compaction, and the absence of function-specific chromatin labels have been the key technical obstacles. The correlation between DNA sequence composition and its contribution to the chromosome-scale structure has been suggested before; it is unclear though if all DNA sequences equally participate in intra- or inter-chromatin or DNA-protein interactions that lead to formation of mitotic chromosomes and if their mitotic positions are reproduced radially. Using high-resolution fluorescence microscopy of live or minimally perturbed, fixed chromosomes in Drosophila embryonic cultures or tissues expressing MSL3-GFP fusion protein, we studied positioning of specific MSL3-binding sites. Actively transcribed, dosage compensated Drosophila genes are distributed along the euchromatic arm of the male X chromosome. Several novel features of mitotic chromosomes have been observed. MSL3-GFP is always found at the periphery of mitotic chromosomes, suggesting that active, dosage compensated genes are also found at the periphery of mitotic chromosomes. Furthermore, radial distribution of chromatin loci on mitotic chromosomes was found to be correlated with their functional activity as judged by core histone modifications. Histone modifications specific to active chromatin were found peripheral with respect to silent chromatin. MSL3-GFP-labeled chromatin loci become peripheral starting in late prophase. In early prophase, dosage compensated chromatin regions traverse the entire width of chromosomes. These findings suggest large-scale internal rearrangements within chromosomes during the prophase condensation step, arguing against consecutive coiling models. Our results suggest that the organization of mitotic chromosomes is reproducible not only longitudinally, as demonstrated by chromosome-specific banding patterns, but also radially. Specific MSL3-binding sites, the majority of which have been demonstrated earlier to be dosage compensated DNA sequences, located on the X chromosomes, and actively transcribed in interphase, are positioned at the periphery of mitotic chromosomes. This potentially describes a connection between the DNA/protein content of chromatin loci and their contribution to mitotic chromosome structure. Live high-resolution observations of consecutive condensation states in MSL3-GFP expressing cells could provide additional details regarding the condensation mechanisms.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Belmont A. S. Mitotic chromosome structure and condensation. Curr Opin Cell Biol. 2006;18:632–638. - PubMed
-
- Dietzel S, Belmont A. S. Reproducible but dynamic positioning of DNA in chromosomes during mitosis. Nat Cell Biol. 2001;3:767–770. - PubMed
-
- Marko J. F. Micromechanical studies of mitotic chromosomes. Chromosome Res. 2008;16:469–497. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

