The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing
- PMID: 21264352
- PMCID: PMC3019111
- DOI: 10.1371/journal.pbio.1000573
The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing
Abstract
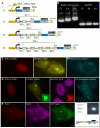
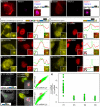
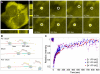
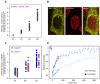
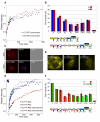
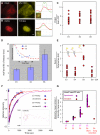
RNA processing events that take place on the transcribed pre-mRNA include capping, splicing, editing, 3' processing, and polyadenylation. Most of these processes occur co-transcriptionally while the RNA polymerase II (Pol II) enzyme is engaged in transcriptional elongation. How Pol II elongation rates are influenced by splicing is not well understood. We generated a family of inducible gene constructs containing increasing numbers of introns and exons, which were stably integrated in human cells to serve as actively transcribing gene loci. By monitoring the association of the transcription and splicing machineries on these genes in vivo, we showed that only U1 snRNP localized to the intronless gene, consistent with a splicing-independent role for U1 snRNP in transcription. In contrast, all snRNPs accumulated on intron-containing genes, and increasing the number of introns increased the amount of spliceosome components recruited. This indicates that nascent RNA can assemble multiple spliceosomes simultaneously. Kinetic measurements of Pol II elongation in vivo, Pol II ChIP, as well as use of Spliceostatin and Meayamycin splicing inhibitors showed that polymerase elongation rates were uncoupled from ongoing splicing. This study shows that transcription elongation kinetics proceed independently of splicing at the model genes studied here. Surprisingly, retention of polyadenylated mRNA was detected at the transcription site after transcription termination. This suggests that the polymerase is released from chromatin prior to the completion of splicing, and the pre-mRNA is post-transcriptionally processed while still tethered to chromatin near the gene end.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Comment in
-
Gene expression: RNAPII stands alone.Nat Rev Mol Cell Biol. 2011 Mar;12(3):136-7. doi: 10.1038/nrm3070. Nat Rev Mol Cell Biol. 2011. PMID: 21346727 No abstract available.
References
-
- Bentley D. L. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. - PubMed
-
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. - PubMed
-
- Moore M. J, Proudfoot N. J. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

