The logic of kinetic regulation in the thioredoxin system
- PMID: 21266044
- PMCID: PMC3045320
- DOI: 10.1186/1752-0509-5-15
The logic of kinetic regulation in the thioredoxin system
Abstract
Background: The thioredoxin system consisting of NADP(H), thioredoxin reductase and thioredoxin provides reducing equivalents to a large and diverse array of cellular processes. Despite a great deal of information on the kinetics of individual thioredoxin-dependent reactions, the kinetic regulation of this system as an integrated whole is not known. We address this by using kinetic modeling to identify and describe kinetic behavioral motifs found within the system.
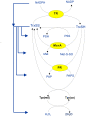
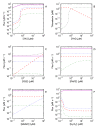
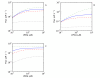
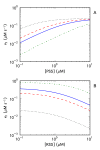
Results: Analysis of a realistic computational model of the Escherichia coli thioredoxin system revealed several modes of kinetic regulation in the system. In keeping with published findings, the model showed that thioredoxin-dependent reactions were adaptable (i.e. changes to the thioredoxin system affected the kinetic profiles of these reactions). Further and in contrast to other systems-level descriptions, analysis of the model showed that apparently unrelated thioredoxin oxidation reactions can affect each other via their combined effects on the thioredoxin redox cycle. However, the scale of these effects depended on the kinetics of the individual thioredoxin oxidation reactions with some reactions more sensitive to changes in the thioredoxin cycle and others, such as the Tpx-dependent reduction of hydrogen peroxide, less sensitive to these changes. The coupling of the thioredoxin and Tpx redox cycles also allowed for ultrasensitive changes in the thioredoxin concentration in response to changes in the thioredoxin reductase concentration. We were able to describe the kinetic mechanisms underlying these behaviors precisely with analytical solutions and core models.
Conclusions: Using kinetic modeling we have revealed the logic that underlies the functional organization and kinetic behavior of the thioredoxin system. The thioredoxin redox cycle and associated reactions allows for a system that is adaptable, interconnected and able to display differential sensitivities to changes in this redox cycle. This work provides a theoretical, systems-biological basis for an experimental analysis of the thioredoxin system and its associated reactions.
Figures

References
-
- Vlamis-Gardikas A. The multiple functions of the thiol-based electron flow pathways of Escherichia coli: Eternal concepts revisited. Biochim Biophys Acta. 2008;1780:1170–1200. - PubMed
-
- Holmgren A, Bjornstedt M. Thioredoxin and thioredoxin reductase. Methods Enzymol. 1995;252:199–208. full_text. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

