Characterisation and expression of microRNAs in developing wings of the neotropical butterfly Heliconius melpomene
- PMID: 21266089
- PMCID: PMC3039609
- DOI: 10.1186/1471-2164-12-62
Characterisation and expression of microRNAs in developing wings of the neotropical butterfly Heliconius melpomene
Abstract
Background: Heliconius butterflies are an excellent system for studies of adaptive convergent and divergent phenotypic traits. Wing colour patterns are used as signals to both predators and potential mates and are inherited in a Mendelian manner. The underlying genetic mechanisms of pattern formation have been studied for many years and shed light on broad issues, such as the repeatability of evolution. In Heliconius melpomene, the yellow hindwing bar is controlled by the HmYb locus. MicroRNAs (miRNAs) are important post-transcriptional regulators of gene expression that have key roles in many biological processes, including development. miRNAs could act as regulators of genes involved in wing development, patterning and pigmentation. For this reason we characterised miRNAs in developing butterfly wings and examined differences in their expression between colour pattern races.
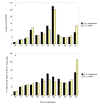
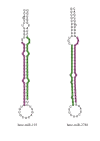
Results: We sequenced small RNA libraries from two colour pattern races and detected 142 Heliconius miRNAs with homology to others found in miRBase. Several highly abundant miRNAs were differentially represented in the libraries between colour pattern races. These candidates were tested further using Northern blots, showing that differences in expression were primarily due to developmental stage rather than colour pattern. Assembly of sequenced reads to the HmYb region identified hme-miR-193 and hme-miR-2788; located 2380 bp apart in an intergenic region. These two miRNAs are expressed in wings and show an upregulation between 24 and 72 hours post-pupation, indicating a potential role in butterfly wing development. A search for miRNAs in all available H. melpomene BAC sequences (~2.5 Mb) did not reveal any other miRNAs and no novel miRNAs were predicted.
Conclusions: Here we describe the first butterfly miRNAs and characterise their expression in developing wings. Some show differences in expression across developing pupal stages and may have important functions in butterfly wing development. Two miRNAs were located in the HmYb region and were expressed in developing pupal wings. Future work will examine the expression of these miRNAs in different colour pattern races and identify miRNA targets among wing patterning genes.
Figures


References
-
- Langham GM. "Specialized Avian Predators Repeatedly Attack Novel Color Morphs of Heliconius Butterflies,". Evolution. 2004;58(12):2783–2787. - PubMed
-
- Sheppard PM, Turner JRG, Brown KS, Benson WW, Singer MC. "Genetics and the Evolution of Muellerian Mimicry in Heliconius Butterflies,". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 1985;308(1137):433–610. doi: 10.1098/rstb.1985.0066. - DOI
-
- Mallet J. "The Genetics of Warning Colour in Peruvian Hybrid Zones of Heliconius erato and H. melpomene,". Proceedings of the Royal Society of London. B. Biological Sciences. 1989;236(1283):163–185. doi: 10.1098/rspb.1989.0019. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

