Identification of evolutionarily conserved non-AUG-initiated N-terminal extensions in human coding sequences
- PMID: 21266472
- PMCID: PMC3105428
- DOI: 10.1093/nar/gkr007
Identification of evolutionarily conserved non-AUG-initiated N-terminal extensions in human coding sequences
Abstract
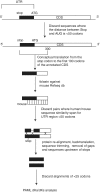
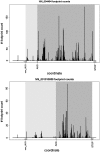
In eukaryotes, it is generally assumed that translation initiation occurs at the AUG codon closest to the messenger RNA 5' cap. However, in certain cases, initiation can occur at codons differing from AUG by a single nucleotide, especially the codons CUG, UUG, GUG, ACG, AUA and AUU. While non-AUG initiation has been experimentally verified for a handful of human genes, the full extent to which this phenomenon is utilized--both for increased coding capacity and potentially also for novel regulatory mechanisms--remains unclear. To address this issue, and hence to improve the quality of existing coding sequence annotations, we developed a methodology based on phylogenetic analysis of predicted 5' untranslated regions from orthologous genes. We use evolutionary signatures of protein-coding sequences as an indicator of translation initiation upstream of annotated coding sequences. Our search identified novel conserved potential non-AUG-initiated N-terminal extensions in 42 human genes including VANGL2, FGFR1, KCNN4, TRPV6, HDGF, CITED2, EIF4G3 and NTF3, and also affirmed the conservation of known non-AUG-initiated extensions in 17 other genes. In several instances, we have been able to obtain independent experimental evidence of the expression of non-AUG-initiated products from the previously published literature and ribosome profiling data.
Figures





References
-
- Ramakrishnan V. Ribosome structure and the mechanism of translation. Cell. 2002;108:557–572. - PubMed
-
- Simonetti A, Marzi S, Myasnikov AG, Fabbretti A, Yusupov M, Gualerzi CO, Klaholz BP. Structure of the 30S translation initiation complex. Nature. 2008;455:416–420. - PubMed
-
- Potapov AP, Triana-Alonso FJ, Nierhaus KH. Ribosomal decoding processes at codons in the A or P sites depend differently on 2′-OH groups. J. Biol. Chem. 1995;270:17680–17684. - PubMed
-
- Ogle JM, Brodersen DE, Clemons WM, Jr, Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

