Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor
- PMID: 21266545
- PMCID: PMC3038704
- DOI: 10.1073/pnas.1019217108
Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor
Abstract
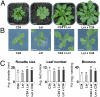
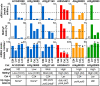
Intraspecific hybrids between the Arabidopsis thaliana accessions C24 and Landsberg erecta have strong heterosis. The reciprocal hybrids show a decreased level of 24-nt small RNA (sRNA) relative to the parents with the decrease greatest for those loci where the parents had markedly different 24-nt sRNA levels. The genomic regions with reduced 24-nt sRNA levels were largely associated with genes and their flanking regions indicating a potential effect on gene expression. We identified several examples of genes with altered 24-nt sRNA levels that showed correlated changes in DNA methylation and expression levels. We suggest that such epigenetically generated differences in gene activity may contribute to hybrid vigor and that the epigenetic diversity between ecotypes provides increased allelic (epi-allelic) variability that could contribute to heterosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
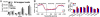



References
-
- Shull GH. Beginnings of the heterosis concept. In: Gowan JW, editor. Heterosis. Ames, IA: Iowa State College Press; 1952. pp. 14–48.
-
- Hochholdinger F, Hoecker N. Towards the molecular basis of heterosis. Trends Plant Sci. 2007;12:427–432. - PubMed
-
- Lippman ZB, Zamir D. Heterosis: Revisiting the magic. Trends Genet. 2007;23:60–66. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

