Genome-wide RNAi screen reveals disease-associated genes that are common to Hedgehog and Wnt signaling
- PMID: 21266715
- PMCID: PMC3790583
- DOI: 10.1126/scisignal.2001225
Genome-wide RNAi screen reveals disease-associated genes that are common to Hedgehog and Wnt signaling
Abstract
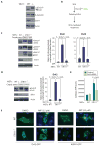
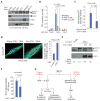
The Hedgehog (Hh) and Wnt signal transduction pathways are master regulators of embryogenesis and tissue renewal and represent anticancer therapeutic targets. Using genome-wide RNA interference screening in murine cultured cells, we established previously unknown associations between these signaling pathways and genes linked to developmental malformations, diseases of premature tissue degeneration, and cancer. We identified functions in both pathways for the multitasking kinase Stk11 (also known as Lkb1), a tumor suppressor implicated in lung and cervical cancers. We found that Stk11 loss resulted in disassembly of the primary cilium, a cellular organizing center for Hh pathway components, thus dampening Hh signaling. Loss of Stk11 also induced aberrant signaling through the Wnt pathway. Chemicals that targeted the Wnt acyltransferase Porcupine or that restored primary cilia length by inhibiting the tubulin deacetylase HDAC6 (histone deacetylase 6) countered deviant pathway activities driven by Stk11 loss. Our study demonstrates that Stk11 is a critical mediator in both the Hh and the Wnt pathways, and our approach provides a platform to support the development of targeted therapeutic strategies.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

