Cryptic prophages help bacteria cope with adverse environments
- PMID: 21266997
- PMCID: PMC3105296
- DOI: 10.1038/ncomms1146
Cryptic prophages help bacteria cope with adverse environments
Abstract
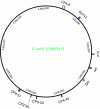
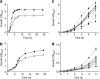
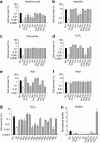
Phages are the most abundant entity in the biosphere and outnumber bacteria by a factor of 10. Phage DNA may also constitute 20% of bacterial genomes; however, its role is ill defined. Here, we explore the impact of cryptic prophages on cell physiology by precisely deleting all nine prophage elements (166 kbp) using Escherichia coli. We find that cryptic prophages contribute significantly to resistance to sub-lethal concentrations of quinolone and β-lactam antibiotics primarily through proteins that inhibit cell division (for example, KilR of rac and DicB of Qin). Moreover, the prophages are beneficial for withstanding osmotic, oxidative and acid stresses, for increasing growth, and for influencing biofilm formation. Prophage CPS-53 proteins YfdK, YfdO and YfdS enhanced resistance to oxidative stress, prophages e14, CPS-53 and CP4-57 increased resistance to acid, and e14 and rac proteins increased early biofilm formation. Therefore, cryptic prophages provide multiple benefits to the host for surviving adverse environmental conditions.
Figures

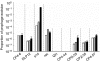
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

