Synthetic associations are unlikely to account for many common disease genome-wide association signals
- PMID: 21267062
- PMCID: PMC3022527
- DOI: 10.1371/journal.pbio.1000580
Synthetic associations are unlikely to account for many common disease genome-wide association signals
Abstract
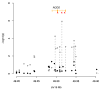
Synthetic associations have been posited as a possible explanation for missing heritability in complex disease. We show several lines of evidence which suggest that, while possible, these synthetic associations are not common.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Comment in
-
The importance of synthetic associations will only be resolved empirically.PLoS Biol. 2011 Jan 18;9(1):e1001008. doi: 10.1371/journal.pbio.1001008. PLoS Biol. 2011. PMID: 21267066 Free PMC article. No abstract available.
Comment on
-
Rare variants create synthetic genome-wide associations.PLoS Biol. 2010 Jan 26;8(1):e1000294. doi: 10.1371/journal.pbio.1000294. PLoS Biol. 2010. PMID: 20126254 Free PMC article.
References
-
- Haines J. L, Hauser M. A, Schmidt S, Scott W. K, Olson L. M, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

