HLTF and SHPRH are not essential for PCNA polyubiquitination, survival and somatic hypermutation: existence of an alternative E3 ligase
- PMID: 21269891
- PMCID: PMC3381732
- DOI: 10.1016/j.dnarep.2010.12.008
HLTF and SHPRH are not essential for PCNA polyubiquitination, survival and somatic hypermutation: existence of an alternative E3 ligase
Abstract
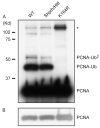
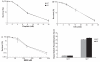
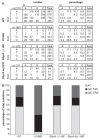
DNA damage tolerance is regulated at least in part at the level of proliferating cell nuclear antigen (PCNA) ubiquitination. Monoubiquitination (PCNA-Ub) at lysine residue 164 (K164) stimulates error-prone translesion synthesis (TLS), Rad5-dependent polyubiquitination (PCNA-Ub(n)) stimulates error-free template switching (TS). To generate high affinity antibodies by somatic hypermutation (SHM), B cells profit from error-prone TLS polymerases. Consistent with the role of PCNA-Ub in stimulating TLS, hypermutated B cells of PCNA(K164R) mutant mice display a defect in generating selective point mutations. Two Rad5 orthologs, HLTF and SHPRH have been identified as alternative E3 ligases generating PCNA-Ub(n) in mammals. As PCNA-Ub and PCNA-Ub(n) both make use of K164, error-free PCNA-Ub(n)-dependent TS may suppress error-prone PCNA-Ub-dependent TLS. To determine a regulatory role of Shprh and Hltf in SHM, we generated Shprh/Hltf double mutant mice. Interestingly, while the formation of PCNA-Ub and PCNA-Ub(n) is prohibited in PCNA(K164R) MEFs, the formation of PCNA-Ub(n) is not abolished in Shprh/Hltf mutant MEFs. In line with these observations Shprh/Hltf double mutant B cells were not hypersensitive to DNA damage. Furthermore, SHM was normal in Shprh/Hltf mutant B cells. These data suggest the existence of an alternative E3 ligase in the generation of PCNA-Ub(n).
Copyright © 2010 Elsevier B.V. All rights reserved.
Figures

Similar articles
-
Helicase-Like Transcription Factor HLTF and E3 Ubiquitin Ligase SHPRH Confer DNA Damage Tolerance through Direct Interactions with Proliferating Cell Nuclear Antigen (PCNA).Int J Mol Sci. 2020 Jan 21;21(3):693. doi: 10.3390/ijms21030693. Int J Mol Sci. 2020. PMID: 31973093 Free PMC article.
-
PCNA ubiquitination-independent activation of polymerase η during somatic hypermutation and DNA damage tolerance.DNA Repair (Amst). 2011 Oct 10;10(10):1051-9. doi: 10.1016/j.dnarep.2011.08.005. Epub 2011 Sep 1. DNA Repair (Amst). 2011. PMID: 21889916
-
Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks.Proc Natl Acad Sci U S A. 2008 Aug 26;105(34):12411-6. doi: 10.1073/pnas.0805685105. Epub 2008 Aug 21. Proc Natl Acad Sci U S A. 2008. PMID: 18719106 Free PMC article.
-
Regulation of DNA damage tolerance in mammalian cells by post-translational modifications of PCNA.Mutat Res. 2017 Oct;803-805:82-88. doi: 10.1016/j.mrfmmm.2017.06.004. Epub 2017 Jun 21. Mutat Res. 2017. PMID: 28666590 Review.
-
Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance.DNA Repair (Amst). 2010 Mar 2;9(3):257-67. doi: 10.1016/j.dnarep.2009.12.013. Epub 2010 Jan 21. DNA Repair (Amst). 2010. PMID: 20096653 Review.
Cited by
-
Maneuvers on PCNA Rings during DNA Replication and Repair.Genes (Basel). 2018 Aug 17;9(8):416. doi: 10.3390/genes9080416. Genes (Basel). 2018. PMID: 30126151 Free PMC article. Review.
-
DNA replication: Mechanisms and therapeutic interventions for diseases.MedComm (2020). 2023 Feb 5;4(1):e210. doi: 10.1002/mco2.210. eCollection 2023 Feb. MedComm (2020). 2023. PMID: 36776764 Free PMC article. Review.
-
Strand invasion by HLTF as a mechanism for template switch in fork rescue.Nucleic Acids Res. 2014 Feb;42(3):1711-20. doi: 10.1093/nar/gkt1040. Epub 2013 Nov 5. Nucleic Acids Res. 2014. PMID: 24198246 Free PMC article.
-
The Human RAD5 Homologs, HLTF and SHPRH, Have Separate Functions in DNA Damage Tolerance Dependent on The DNA Lesion Type.Biomolecules. 2020 Mar 17;10(3):463. doi: 10.3390/biom10030463. Biomolecules. 2020. PMID: 32192191 Free PMC article.
-
Two replication fork maintenance pathways fuse inverted repeats to rearrange chromosomes.Nature. 2013 Sep 26;501(7468):569-72. doi: 10.1038/nature12500. Epub 2013 Sep 8. Nature. 2013. PMID: 24013173 Free PMC article.
References
-
- Friedberg EC. Suffering in silence: the tolerance of DNA damage. Nat. Rev. Mol. Cell Biol. 2005;6:943–953. - PubMed
-
- Jansen JG, Fousteri MI, de WN. Send in the clamps: control of DNA translesion synthesis in eukaryotes. Mol. Cell. 2007;28:522–529. - PubMed
-
- Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. - PubMed
-
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. - PubMed
-
- Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

