Capicua DNA-binding sites are general response elements for RTK signaling in Drosophila
- PMID: 21270056
- PMCID: PMC3035094
- DOI: 10.1242/dev.057729
Capicua DNA-binding sites are general response elements for RTK signaling in Drosophila
Abstract
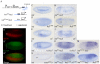
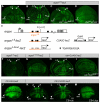
RTK/Ras/MAPK signaling pathways play key functions in metazoan development, but how they control expression of downstream genes is not well understood. In Drosophila, it is generally assumed that most transcriptional responses to RTK signal activation depend on binding of Ets-family proteins to specific cis-acting sites in target enhancers. Here, we show that several Drosophila RTK pathways control expression of downstream genes through common octameric elements that are binding sites for the HMG-box factor Capicua, a transcriptional repressor that is downregulated by RTK signaling in different contexts. We show that Torso RTK-dependent regulation of terminal gap gene expression in the early embryo critically depends on Capicua octameric sites, and that binding of Capicua to these sites is essential for recruitment of the Groucho co-repressor to the huckebein enhancer in vivo. We then show that subsequent activation of the EGFR RTK pathway in the neuroectodermal region of the embryo controls dorsal-ventral gene expression by downregulating the Capicua protein, and that this control also depends on Capicua octameric motifs. Thus, a similar mechanism of RTK regulation operates during subdivision of the anterior-posterior and dorsal-ventral embryonic axes. We also find that identical DNA octamers mediate Capicua-dependent regulation of another EGFR target in the developing wing. Remarkably, a simple combination of activator-binding sites and Capicua motifs is sufficient to establish complex patterns of gene expression in response to both Torso and EGFR activation in different tissues. We conclude that Capicua octamers are general response elements for RTK signaling in Drosophila.
Figures




References
-
- Barolo S., Posakony J. W. (2002). Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 16, 1167-1181 - PubMed
-
- Blair S. S. (2007). Wing vein patterning in Drosophila and the analysis of intercellular signaling. Annu. Rev. Cell Dev. Biol. 23, 293-319 - PubMed
-
- Brönner G., Jäckle H. (1991). Control and function of terminal gap gene activity in the posterior pole region of the Drosophila embryo. Mech. Dev. 35, 205-211 - PubMed
-
- Chen Y. J., Chiang C. S., Weng L. C., Lengyel J. A., Liaw G. J. (2002). Tramtrack69 is required for the early repression of tailless expression. Mech. Dev. 116, 75-83 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

