Viral metagenome analysis to guide human pathogen monitoring in environmental samples
- PMID: 21272046
- PMCID: PMC3055918
- DOI: 10.1111/j.1472-765X.2011.03014.x
Viral metagenome analysis to guide human pathogen monitoring in environmental samples
Abstract
Aims: The aim of this study was to develop and demonstrate an approach for describing the diversity of human pathogenic viruses in an environmentally isolated viral metagenome.
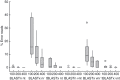
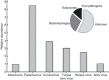
Methods and results: In silico bioinformatic experiments were used to select an optimum annotation strategy for discovering human viruses in virome data sets and applied to annotate a class B biosolid virome. Results from the in silico study indicated that <1% errors in virus identification could be achieved when nucleotide-based search programs (BLASTn or tBLASTx), viral genome only databases and sequence reads >200 nt were considered. Within the 51,925 annotated sequences, 94 DNA and 19 RNA sequences were identified as human viruses. Virus diversity included environmentally transmitted agents such as parechovirus, coronavirus, adenovirus and aichi virus, as well as viruses associated with chronic human infections such as human herpes and hepatitis C viruses.
Conclusions: This study provided a bioinformatic approach for identifying pathogens in a virome data set and demonstrated the human virus diversity in a relevant environmental sample.
Significance and impact of the study: As the costs of next-generation sequencing decrease, the pathogen diversity described by virus metagenomes will provide an unbiased guide for subsequent cell culture and quantitative pathogen analyses and ensures that highly enriched and relevant pathogens are not neglected in exposure and risk assessments.
© 2011 The Authors. Letters in Applied Microbiology © 2011 The Society for Applied Microbiology.
Figures
References
-
- Baumgarte,S., de Souza Luna,L.K., Grywna,K., Panning,M., Drexler,J.F., Karsten,C., Huppertz,H.I. and Drosten,C. (2008) Prevalence, types, and RNA concentrations of human parechoviruses, including a sixth parechovirus type, in stool samples from patients with acute enteritis. J Clin Microbiol 46, 242–248. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

