Comparative genomics allowed the identification of drug targets against human fungal pathogens
- PMID: 21272313
- PMCID: PMC3042012
- DOI: 10.1186/1471-2164-12-75
Comparative genomics allowed the identification of drug targets against human fungal pathogens
Abstract
Background: The prevalence of invasive fungal infections (IFIs) has increased steadily worldwide in the last few decades. Particularly, there has been a global rise in the number of infections among immunosuppressed people. These patients present severe clinical forms of the infections, which are commonly fatal, and they are more susceptible to opportunistic fungal infections than non-immunocompromised people. IFIs have historically been associated with high morbidity and mortality, partly because of the limitations of available antifungal therapies, including side effects, toxicities, drug interactions and antifungal resistance. Thus, the search for alternative therapies and/or the development of more specific drugs is a challenge that needs to be met. Genomics has created new ways of examining genes, which open new strategies for drug development and control of human diseases.
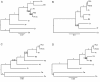
Results: In silico analyses and manual mining selected initially 57 potential drug targets, based on 55 genes experimentally confirmed as essential for Candida albicans or Aspergillus fumigatus and other 2 genes (kre2 and erg6) relevant for fungal survival within the host. Orthologs for those 57 potential targets were also identified in eight human fungal pathogens (C. albicans, A. fumigatus, Blastomyces dermatitidis, Paracoccidioides brasiliensis, Paracoccidioides lutzii, Coccidioides immitis, Cryptococcus neoformans and Histoplasma capsulatum). Of those, 10 genes were present in all pathogenic fungi analyzed and absent in the human genome. We focused on four candidates: trr1 that encodes for thioredoxin reductase, rim8 that encodes for a protein involved in the proteolytic activation of a transcriptional factor in response to alkaline pH, kre2 that encodes for α-1,2-mannosyltransferase and erg6 that encodes for Δ(24)-sterol C-methyltransferase.
Conclusions: Our data show that the comparative genomics analysis of eight fungal pathogens enabled the identification of four new potential drug targets. The preferred profile for fungal targets includes proteins conserved among fungi, but absent in the human genome. These characteristics potentially minimize toxic side effects exerted by pharmacological inhibition of the cellular targets. From this first step of post-genomic analysis, we obtained information relevant to future new drug development.
Figures
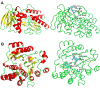
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

