Eukaryote-wide sequence analysis of mitochondrial β-barrel outer membrane proteins
- PMID: 21272379
- PMCID: PMC3045335
- DOI: 10.1186/1471-2164-12-79
Eukaryote-wide sequence analysis of mitochondrial β-barrel outer membrane proteins
Abstract
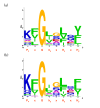
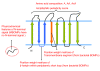
Background: The outer membranes of mitochondria are thought to be homologous to the outer membranes of Gram negative bacteria, which contain 100's of distinct families of β-barrel membrane proteins (BOMPs) often forming channels for transport of nutrients or drugs. However, only four families of mitochondrial BOMPs (MBOMPs) have been confirmed to date. Although estimates as high as 100 have been made in the past, the number of yet undiscovered MBOMPs is an open question. Fortunately, the recent discovery of a membrane integration signal (the β-signal) for MBOMPs gave us an opportunity to look for undiscovered MBOMPs.
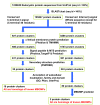
Results: We present the results of a comprehensive survey of eukaryotic protein sequences intended to identify new MBOMPs. Our search employs recent results on β-signals as well as structural information and a novel BOMP predictor trained on both bacterial and mitochondrial BOMPs. Our principal finding is circumstantial evidence suggesting that few MBOMPs remain to be discovered, if one assumes that, like known MBOMPs, novel MBOMPs will be monomeric and β-signal dependent. In addition to this, our analysis of MBOMP homologs reveals some exceptions to the current model of the β-signal, but confirms its consistent presence in the C-terminal region of MBOMP proteins. We also report a β-signal independent search for MBOMPs against the yeast and Arabidopsis proteomes. We find no good candidates MBOMPs in yeast but the Arabidopsis results are less conclusive.
Conclusions: Our results suggest there are no remaining MBOMPs left to discover in yeast; and if one assumes all MBOMPs are β-signal dependent, few MBOMP families remain undiscovered in any sequenced organism.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

