Origin, functional role, and clinical impact of Fanconi anemia FANCA mutations
- PMID: 21273304
- PMCID: PMC3083295
- DOI: 10.1182/blood-2010-08-299917
Origin, functional role, and clinical impact of Fanconi anemia FANCA mutations
Abstract
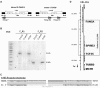
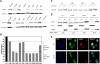
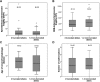
Fanconi anemia is characterized by congenital abnormalities, bone marrow failure, and cancer predisposition. To investigate the origin, functional role, and clinical impact of FANCA mutations, we determined a FANCA mutational spectrum with 130 pathogenic alleles. Some of these mutations were further characterized for their distribution in populations, mode of emergence, or functional consequences at cellular and clinical level. The world most frequent FANCA mutation is not the result of a mutational "hot-spot" but results from worldwide dissemination of an ancestral Indo-European mutation. We provide molecular evidence that total absence of FANCA in humans does not reduce embryonic viability, as the observed frequency of mutation carriers in the Gypsy population equals the expected by Hardy-Weinberg equilibrium. We also prove that long distance Alu-Alu recombination can cause Fanconi anemia by originating large interstitial deletions involving FANCA and 2 adjacent genes. Finally, we show that all missense mutations studied lead to an altered FANCA protein that is unable to relocate to the nucleus and activate the FA/BRCA pathway. This may explain the observed lack of correlation between type of FANCA mutation and cellular phenotype or clinical severity in terms of age of onset of hematologic disease or number of malformations.
Figures



References
-
- Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8(10):735–748. - PubMed
-
- Meindl A, Hellebrand H, Wiek C, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet. 2010;42(5):410–414. - PubMed
-
- Joenje H, Patel KJ. The emerging genetic and molecular basis of Fanconi anaemia. Nat Rev Genet. 2001;2(6):446–457. - PubMed
-
- Kennedy RD, D'Andrea AD. The Fanconi anemia/BRCA pathway: new faces in the crowd. Genes Dev. 2005;19(24):2925–2940. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

