Tuning the period of the mammalian circadian clock: additive and independent effects of CK1εTau and Fbxl3Afh mutations on mouse circadian behavior and molecular pacemaking
- PMID: 21273438
- PMCID: PMC6623629
- DOI: 10.1523/JNEUROSCI.4107-10.2011
Tuning the period of the mammalian circadian clock: additive and independent effects of CK1εTau and Fbxl3Afh mutations on mouse circadian behavior and molecular pacemaking
Abstract
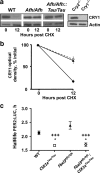
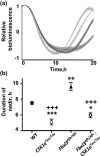
Circadian pacemaking in the suprachiasmatic nucleus (SCN) revolves around a transcriptional/posttranslational feedback loop in which period (Per) and cryptochrome (Cry) genes are negatively regulated by their protein products. Genetically specified differences in this oscillator underlie sleep and metabolic disorders, and dictate diurnal/nocturnal preference. A critical goal, therefore, is to identify mechanisms that generate circadian phenotypic diversity, through both single gene effects and gene interactions. The individual stabilities of PER or CRY proteins determine pacemaker period, and PER/CRY complexes have been proposed to afford mutual stabilization, although how PER and CRY proteins with contrasting stabilities interact is unknown. We therefore examined interactions between two mutations in male mice: Fbxl3(Afh), which lengthens period by stabilizing CRY, and Csnk1ε(tm1Asil) (CK1ε(Tau)), which destabilizes PER, thereby accelerating the clock. By intercrossing these mutants, we show that the stabilities of CRY and PER are independently regulated, contrary to the expectation of mutual stabilization. Segregation of wild-type and mutant alleles generated a spectrum of periods for rest-activity behavior and SCN bioluminescence rhythms. The mutations exerted independent, additive effects on circadian period, biased toward shorter periods determined by CK1ε(Tau). Notably, Fbxl3(Afh) extended the duration of the nadir of the PER2-driven bioluminescence rhythm but CK1ε(Tau) reversed this, indicating that despite maintained CRY expression, CK1ε(Tau) truncated the interval of negative feedback. These results argue for independent, additive biochemical actions of PER and CRY in circadian control, and complement genome-wide epistatic analyses, seeking to decipher the multigenic control of circadian pacemaking.
Figures


References
-
- Allebrandt KV, Teder-Laving M, Akyol M, Pichler I, Müller-Myhsok B, Pramstaller P, Merrow M, Meitinger T, Metspalu A, Roenneberg T. CLOCK gene variants associate with sleep duration in two independent populations. Biol Psychiatry. 2010;67:1040–1047. - PubMed
-
- Dey J, Carr AJ, Cagampang FR, Semikhodskii AS, Loudon AS, Hastings MH, Maywood ES. The tau mutation in the Syrian hamster differentially reprograms the circadian clock in the SCN and peripheral tissues. J Biol Rhythms. 2005;20:99–110. - PubMed
-
- Godinho SI, Maywood ES, Shaw L, Tucci V, Barnard AR, Busino L, Pagano M, Kendall R, Quwailid MM, Romero MR, O'Neill J, Chesham JE, Brooker D, Lalanne Z, Hastings MH, Nolan PM. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897–900. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/E022553/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/E023223/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MC_U105170643/MRC_/Medical Research Council/United Kingdom
- MC_U142684173/MRC_/Medical Research Council/United Kingdom
- BBE0225531/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
