Ligand-dependent degradation of SRC-1 is pivotal for progesterone receptor transcriptional activity
- PMID: 21273440
- PMCID: PMC3320859
- DOI: 10.1210/me.2010-0458
Ligand-dependent degradation of SRC-1 is pivotal for progesterone receptor transcriptional activity
Abstract
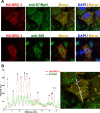
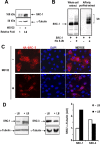
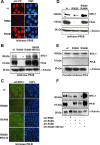
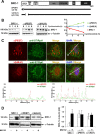
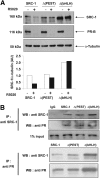
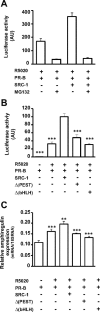
The progesterone receptor (PR), a ligand-activated transcription factor, recruits the primary coactivator steroid receptor coactivator-1 (SRC-1) gene promoters. It is known that PR transcriptional activity is paradoxically coupled to its ligand-dependent down-regulation. However, despite its importance in PR function, the regulation of SRC-1 expression level during hormonal exposure is poorly understood. Here we report that SRC-1 expression level (but not other p160 family members) is down-regulated by the agonist ligand R5020 in a PR-dependent manner. In contrast, the antagonist RU486 fails to induce down-regulation of the coactivator and impairs PR agonist-dependent degradation of SRC-1. We show that SRC-1 proteolysis is a proteasome- and ubiquitin-mediated process that, predominantly but not exclusively, occurs in the cytoplasmic compartment in which SRC-1 colocalizes with proteasome antigens as demonstrated by confocal imaging. Moreover, SRC-1 was stabilized in the presence of leptomycin B or several proteasomal inhibitors. Two degradation motifs, amino-acids 2-16 corresponding to a PEST motif and amino acids 41-136 located in the basic helix loop helix domain of the coactivator, were identified and shown to control the stability as well as the hormone-dependent down-regulation of the coactivator. SRC-1 degradation is of physiological importance because the two nondegradable mutants that still interacted with PR as demonstrated by coimmunoprecipitation failed to stimulate transcription of exogenous and endogenous target genes, suggesting that concomitant PR/SRC-1 ligand-dependent degradation is a necessary step for PR transactivation activity. Collectively our findings are consistent with the emerging role of proteasome-mediated proteolysis in the gene-regulating process and indicate that the ligand-dependent down-regulation of SRC-1 is critical for PR transcriptional activity.
Figures

References
-
- Li X , O'Malley BW. 2003. Unfolding the action of progesterone receptors. J Biol Chem 278:39261–39264 - PubMed
-
- Lonard DM , O'Malley BW. 2006. The expanding cosmos of nuclear receptor coactivators. Cell 125:411–414 - PubMed
-
- Xu J , Li Q. 2003. Review of the in vivo functions of the p160 steroid receptor coactivator family. Mol Endocrinol 17:1681–1692 - PubMed
-
- Chen D , Ma H , Hong H , Koh SS , Huang SM , Schurter BT , Aswad DW , Stallcup MR. 1999. Regulation of transcription by a protein methyltransferase. Science 284:2174–2177 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

