S-adenosylmethionine prevents the up regulation of Toll-like receptor (TLR) signaling caused by chronic ethanol feeding in rats
- PMID: 21276439
- PMCID: PMC3092000
- DOI: 10.1016/j.yexmp.2011.01.005
S-adenosylmethionine prevents the up regulation of Toll-like receptor (TLR) signaling caused by chronic ethanol feeding in rats
Abstract
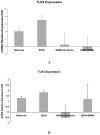
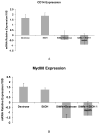
Toll-like receptors (TLR) play a role in mediating the proinflammatory response, fibrogenesis and carcinogenesis in chronic liver diseases such as alcoholic liver disease, non-alcoholic liver disease, hepatitis C and hepatocellular carcinoma. This is true in experimental models of these diseases. For this reason, we investigated the TLR proinflammatory response in the chronic intragastric tube feeding rat model of alcohol liver disease. The methyl donor S-adenosylmethionine was also fed to prevent the gene expression changes induced by ethanol. Ethanol feeding tended to increase the up regulation of the gene expression of TLR2 and TLR4. SAMe feeding prevented this. TLR4 and MyD88 protein levels were significantly increased by ethanol and this was prevented by SAMe. This is the first report where ethanol feeding induced TLR2 and SAMe prevented the induction by ethanol. CD34, FOS, interferon responsive factor 1 (IRF-1), Jun, TLR 1,2,3,4,6 and 7 and Traf-6 were found to be up regulated as seen by microarray analysis where rats were sacrificed at high blood alcohol levels compared to pair fed controls. Il-6, IL-10 and IFNγ were also up regulated by high blood levels of ethanol. The gene expression of CD14, MyD88 and TNFR1SF1 were not up regulated by ethanol but were down regulated by SAMe. The gene expression of IL-1R1 and IRF1 tended to be up regulated by ethanol and this was prevented by feeding SAMe. The results suggest that SAMe, fed chronically prevents the activation of TLR pathways caused by ethanol. In this way the proinflammatory response, fibrogenesis, cirrhosis and hepatocellular carcinoma formation due to alcohol liver disease could be prevented by SAMe.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures




References
-
- Bardag-Gorce F, et al. Gene expression patterns of the liver in response to alcohol: in vivo and in vitro models compared. Exp Mol Pathol. 2006;80:241–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

