Biocompatible quantum dots for biological applications
- PMID: 21276935
- PMCID: PMC3752999
- DOI: 10.1016/j.chembiol.2010.11.013
Biocompatible quantum dots for biological applications
Abstract
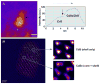
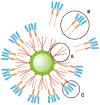
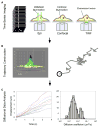
Semiconductor quantum dots are quickly becoming a critical diagnostic tool for discerning cellular function at the molecular level. Their high brightness, long-lasting, size-tunable, and narrow luminescence set them apart from conventional fluorescence dyes. Quantum dots are being developed for a variety of biologically oriented applications, including fluorescent assays for drug discovery, disease detection, single protein tracking, and intracellular reporting. This review introduces the science behind quantum dots and describes how they are made biologically compatible. Several applications are also included, illustrating strategies toward target specificity, and are followed by a discussion on the limitations of quantum dot approaches. The article is concluded with a look at the future direction of quantum dots.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures

References
-
- Alivisatos AP. Perspectives on the physical chemistry of semiconductor nanocrystals. J Phys Chem. 1996a;100:13226–13239.
-
- Alivisatos AP. Semiconductor clusters, nanocrystals, and quantum dots. Science. 1996b;271:933–937.
-
- An S, Zenisek D. Regulation of exocytosis in neurons and neuroendocrine cells. Curr Opin Neurobiol. 2004;14:522–530. - PubMed
-
- Bakalova R, Zhelev Z, Ohba H, Baba Y. Quantum dot-based western blot technology for ultrasensitive detection of tracer proteins. J Am Chem Soc. 2005;127:9328–9329. - PubMed
-
- Ballou B, Lagerholm BC, Ernst LA, Bruchez MP, Waggoner AS. Noninvasive imaging of quantum dots in mice. Bioconjug Chem. 2003;15:79–86. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

