Peptide length and leaving-group sterics influence potency of peptide phosphonate protease inhibitors
- PMID: 21276938
- PMCID: PMC3074588
- DOI: 10.1016/j.chembiol.2010.11.007
Peptide length and leaving-group sterics influence potency of peptide phosphonate protease inhibitors
Abstract
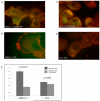
The ability to follow enzyme activity in a cellular context represents a challenging technological frontier that impacts fields ranging from disease pathogenesis to epigenetics. Activity-based probes (ABPs) label the active form of an enzyme via covalent modification of catalytic residues. Here we present an analysis of parameters influencing potency of peptide phosphonate ABPs for trypsin-fold S1A proteases, an abundant and important class of enzymes with similar substrate specificities. We find that peptide length and stability influence potency more than sequence composition and present structural evidence that steric interactions at the prime-side of the substrate-binding cleft affect potency in a protease-dependent manner. We introduce guidelines for the design of peptide phosphonate ABPs and demonstrate their utility in a live-cell labeling application that specifically targets active S1A proteases at the cell surface of cancer cells.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
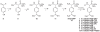

References
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. - PubMed
-
- Behrendt N. The urokinase receptor (uPAR) and the uPAR-associated protein (uPARAP/Endo180): membrane proteins engaged in matrix turnover during tissue remodeling. Biol Chem. 2004;385:103–136. - PubMed
-
- Blum G, Mullins SR, Keren K, Fonovic M, Jedeszko C, Rice MJ, Sloane BF, Bogyo M. Dynamic imaging of protease activity with fluorescently quenched activity-based probes. Nat Chem Biol. 2005;1:203–209. - PubMed
-
- Boduszek B, Oleksyszyn J, Kam CM, Selzler J, Smith RE, Powers JC. Dipeptide phosphonates as inhibitors of dipeptidyl peptidase IV. J Med Chem. 1994;37:3969–3976. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

