Regulation and flexibility of genomic imprinting during seed development
- PMID: 21278124
- PMCID: PMC3051244
- DOI: 10.1105/tpc.110.081018
Regulation and flexibility of genomic imprinting during seed development
Abstract
Genomic imprinting results in monoallelic gene expression in a parent-of-origin-dependent manner. It is achieved by the differential epigenetic marking of parental alleles. Over the past decade, studies in the model systems Arabidopsis thaliana and maize (Zea mays) have shown a strong correlation between silent or active states with epigenetic marks, such as DNA methylation and histone modifications, but the nature of the primary imprint has not been clearly established for all imprinted genes. Phenotypes and expression patterns of imprinted genes have fueled the perception that genomic imprinting is specific to the endosperm, a seed tissue that does not contribute to the next generation. However, several lines of evidence suggest a potential role for imprinting in the embryo, raising questions as to how imprints are erased and reset from one generation to the next. Imprinting regulation in flowering plants shows striking similarities, but also some important differences, compared with the mechanisms of imprinting described in mammals. For example, some imprinted genes are involved in seed growth and viability in plants, which is similar in mammals, where imprinted gene regulation is essential for embryonic development. However, it seems to be more flexible in plants, as imprinting requirements can be bypassed to allow the development of clonal offspring in apomicts.
Figures
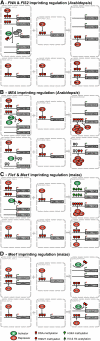
References
-
- Barlow D.P. (1993). Methylation and imprinting: from host defense to gene regulation? Science 260: 309–310 - PubMed
-
- Barlow D.P., Stöger R., Herrmann B.G., Saito K., Schweifer N. (1991). The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature 349: 84–87 - PubMed
-
- Baroux C., Spillane C., Grossniklaus U. (2002). Genomic imprinting during seed development. Adv. Genet. 46: 165–214 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

