Okazaki fragment maturation: nucleases take centre stage
- PMID: 21278448
- PMCID: PMC3030970
- DOI: 10.1093/jmcb/mjq048
Okazaki fragment maturation: nucleases take centre stage
Abstract
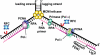
Completion of lagging strand DNA synthesis requires processing of up to 50 million Okazaki fragments per cell cycle in mammalian cells. Even in yeast, the Okazaki fragment maturation happens approximately a million times during a single round of DNA replication. Therefore, efficient processing of Okazaki fragments is vital for DNA replication and cell proliferation. During this process, primase-synthesized RNA/DNA primers are removed, and Okazaki fragments are joined into an intact lagging strand DNA. The processing of RNA/DNA primers requires a group of structure-specific nucleases typified by flap endonuclease 1 (FEN1). Here, we summarize the distinct roles of these nucleases in different pathways for removal of RNA/DNA primers. Recent findings reveal that Okazaki fragment maturation is highly coordinated. The dynamic interactions of polymerase δ, FEN1 and DNA ligase I with proliferating cell nuclear antigen allow these enzymes to act sequentially during Okazaki fragment maturation. Such protein-protein interactions may be regulated by post-translational modifications. We also discuss studies using mutant mouse models that suggest two distinct cancer etiological mechanisms arising from defects in different steps of Okazaki fragment maturation. Mutations that affect the efficiency of RNA primer removal may result in accumulation of unligated nicks and DNA double-strand breaks. These DNA strand breaks can cause varying forms of chromosome aberrations, contributing to development of cancer that associates with aneuploidy and gross chromosomal rearrangement. On the other hand, mutations that impair editing out of polymerase α incorporation errors result in cancer displaying a strong mutator phenotype.
Figures
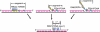

References
-
- Bae S.H., Seo Y.S. Characterization of the enzymatic properties of the yeast Dna2 helicase/endonuclease suggests a new model for Okazaki fragment processing. J. Biol. Chem. 2000;275:38022–38031. doi:10.1074/jbc.M006513200. - DOI - PubMed
-
- Bae S.H., Choi E., Lee K.H., et al. Dna2 of Saccharomyces cerevisiae possesses a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J. Biol. Chem. 1998;273:26880–26890. doi:10.1074/jbc.273.41.26880. - DOI - PubMed
-
- Bae S.H., Bae K.H., Kim J.A., et al. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature. 2001;412:456–461. doi:10.1038/35086609. - DOI - PubMed
-
- Bambara R.A., Murante R.S., Henricksen L.A. Enzymes and reactions at the eukaryotic DNA replication fork. J. Biol. Chem. 1997;272:4647–4650. doi:10.1074/jbc.272.8.4647. - DOI - PubMed
-
- Bentley D.J., Harrison C., Ketchen A.M., et al. DNA ligase I null mouse cells show normal DNA repair activity but altered DNA replication and reduced genome stability. J. Cell Sci. 2002;115:1551–1561. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

