Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5
- PMID: 21278738
- PMCID: PMC3182404
- DOI: 10.1038/ni.1995
Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5
Abstract
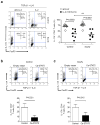
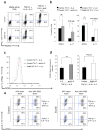
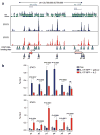
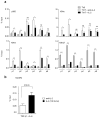
Interleukin 2 (IL-2), a cytokine linked to human autoimmune disease, limits IL-17 production. Here we found that deletion of the gene encoding the transcription factor STAT3 in T cells abrogated IL-17 production and attenuated autoimmunity associated with IL-2 deficiency. Whereas STAT3 induced IL-17 and the transcription factor RORγt and inhibited the transcription factor Foxp3, IL-2 inhibited IL-17 independently of Foxp3 and RORγt. STAT3 and STAT5 bound to multiple common sites across the locus encoding IL-17. The induction of STAT5 binding by IL-2 was associated with less binding of STAT3 at these sites and the inhibition of associated active epigenetic marks. 'Titration' of the relative activation of STAT3 and STAT5 modulated the specification of cells to the IL-17-producing helper T cell (T(H)17 cell) subset. Thus, the balance rather than the absolute magnitude of these signals determined the propensity of cells to make a key inflammatory cytokine.
Conflict of interest statement
Figures

References
-
- Weaver CT, Hatton RD. Interplay between the TH17 and TReg cell lineages: a (co-)evolutionary perspective. Nat Rev Immunol. 2009;9:883–889. - PubMed
-
- Hsu HC, et al. Interleukin 17-producing T helper cells and interleukin 17 orchestrate autoreactive germinal center development in autoimmune BXD2 mice. Nat Immunol. 2008;9:166–175. - PubMed
-
- Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

