Defining the geometry of the two-component proteasome degron
- PMID: 21278740
- PMCID: PMC3129032
- DOI: 10.1038/nchembio.521
Defining the geometry of the two-component proteasome degron
Abstract
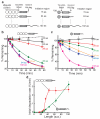
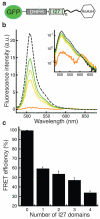
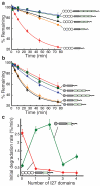
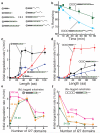
The eukaryotic 26S proteasome controls cellular processes by degrading specific regulatory proteins. Most proteins are targeted for degradation by a signal or degron that consists of two parts: a proteasome-binding tag, typically covalently attached polyubiquitin chains, and an unstructured region that serves as the initiation region for proteasomal proteolysis. Here we have characterized how the arrangement of the two degron parts in a protein affects degradation. We found that a substrate is degraded efficiently only when its initiation region is of a certain minimal length and is appropriately separated in space from the proteasome-binding tag. Regions that are located too close or too far from the proteasome-binding tag cannot access the proteasome and induce degradation. These spacing requirements are different for a polyubiquitin chain and a ubiquitin-like domain. Thus, the arrangement and location of the proteasome initiation region affect a protein's fate and are important in selecting proteins for proteasome-mediated degradation.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

