SORCS1 alters amyloid precursor protein processing and variants may increase Alzheimer's disease risk
- PMID: 21280075
- PMCID: PMC3086759
- DOI: 10.1002/ana.22308
SORCS1 alters amyloid precursor protein processing and variants may increase Alzheimer's disease risk
Abstract
Objective: Sorting mechanisms that cause the amyloid precursor protein (APP) and the β-secretases and γ-secretases to colocalize in the same compartment play an important role in the regulation of Aβ production in Alzheimer's disease (AD). We and others have reported that genetic variants in the Sortilin-related receptor (SORL1) increased the risk of AD, that SORL1 is involved in trafficking of APP, and that underexpression of SORL1 leads to overproduction of Aβ. Here we explored the role of one of its homologs, the sortilin-related VPS10 domain containing receptor 1 (SORCS1), in AD.
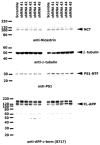
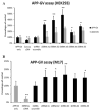
Methods: We analyzed the genetic associations between AD and 16 SORCS1-single nucleotide polymorphisms (SNPs) in 6 independent data sets (2,809 cases and 3,482 controls). In addition, we compared SorCS1 expression levels of affected and unaffected brain regions in AD and control brains in microarray gene expression and real-time polymerase chain reaction (RT-PCR) sets, explored the effects of significant SORCS1-SNPs on SorCS1 brain expression levels, and explored the effect of suppression and overexpression of the common SorCS1 isoforms on APP processing and Aβ generation.
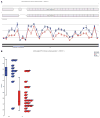
Results: Inherited variants in SORCS1 were associated with AD in all datasets (0.001 < p < 0.049). In addition, SorCS1 influenced APP processing. While overexpression of SorCS1 reduced γ-secretase activity and Aβ levels, the suppression of SorCS1 increased γ-secretase processing of APP and the levels of Aβ.
Interpretations: These data suggest that inherited or acquired changes in SORCS1 expression or function may play a role in the pathogenesis of AD.
Copyright © 2010 American Neurological Association.
Conflict of interest statement
C.C. received grants from The Rosalinde and Arthur Gilbert Foundation/AFAR New Investigator Award in Alzheimer’s Disease, The Taub Institute for Research on Alzheimer’s disease and the Aging Brain, and Columbia University. L.A.F. received grants from an anonymous foundation and the National Institutes of Health, National Institute on Aging. L-N.H. received grants from the Alzheimer Society of Canada, the Canadian Institutes of Health Research (CIHR), Alzheimer Society of Ontario, Howard Hughes Medical Institute, The Wellcome Trust, and the Ontario Research Fund. The laboratory under the direction of P.StG-H. received support from the Alzheimer Society of Canada, CIHR, Alzheimer Society of Ontario, Howard Hughes Medical Institute, The Wellcome Trust, and the Ontario Research Fund. R.M. received grants from the National Institutes of Health, National Institute on Aging, the Blanchette Hooker Rockefeller Foundation, the Charles S. Robertson Gift from the Banbury Fund, and the Merrill Lynch Foundation. A.P. received a grant from
Figures





Comment in
-
Targeting amyloid precursor protein.Ann Neurol. 2011 Jan;69(1):8-10. doi: 10.1002/ana.22342. Ann Neurol. 2011. PMID: 21280070 No abstract available.
References
-
- Edbauer D, Winkler E, Regula JT, et al. Reconstitution of gamma-secretase activity. Nat Cell Biol. 2003;5:486–488. - PubMed
-
- Goate A, Chartier-Harlin MC, Mullan M, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349:704–706. - PubMed
-
- Sherrington R, Rogaev EI, Liang Y, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature. 1995;375:754–760. - PubMed
-
- Rogaev EI, Sherrington R, Rogaeva EA, et al. Familial Alzheimer’s disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer’s disease type 3 gene. Nature. 1995;376:775–778. - PubMed
-
- Harter C, Reinhard C. The secretory pathway from history to the state of the art. Subcell Biochem. 2000;34:1–38. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01-AG09029/AG/NIA NIH HHS/United States
- R01-AG027944/AG/NIA NIH HHS/United States
- CAPMC/ CIHR/Canada
- P01-AG07232/AG/NIA NIH HHS/United States
- P01 AG007232/AG/NIA NIH HHS/United States
- 089703/WT_/Wellcome Trust/United Kingdom
- R01 AG015473/AG/NIA NIH HHS/United States
- R01-AG025259/AG/NIA NIH HHS/United States
- K23AG034550/AG/NIA NIH HHS/United States
- R01-AG019757/AG/NIA NIH HHS/United States
- R37 AG015473/AG/NIA NIH HHS/United States
- R37-AG15473/AG/NIA NIH HHS/United States
- R01 AG041797/AG/NIA NIH HHS/United States
- R01 AG037212/AG/NIA NIH HHS/United States
- R01 AG025259/AG/NIA NIH HHS/United States
- MC_G1000734/MRC_/Medical Research Council/United Kingdom
- R01 AG009029/AG/NIA NIH HHS/United States
- U24 AG021886/AG/NIA NIH HHS/United States
- K23 AG034550/AG/NIA NIH HHS/United States
- P30 AG013846/AG/NIA NIH HHS/United States
- U24 AG056270/AG/NIA NIH HHS/United States
- 081864/WT_/Wellcome Trust/United Kingdom
- P30-AG13846)/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

