An evolved aminoacyl-tRNA synthetase with atypical polysubstrate specificity
- PMID: 21280675
- PMCID: PMC3694404
- DOI: 10.1021/bi101929e
An evolved aminoacyl-tRNA synthetase with atypical polysubstrate specificity
Abstract
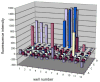
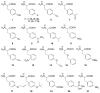
We have employed a rapid fluorescence-based screen to assess the polyspecificity of several aminoacyl-tRNA synthetases (aaRSs) against an array of unnatural amino acids. We discovered that a p-cyanophenylalanine specific aminoacyl-tRNA synthetase (pCNF-RS) has high substrate permissivity for unnatural amino acids, while maintaining its ability to discriminate against the 20 canonical amino acids. This orthogonal pCNF-RS, together with its cognate amber nonsense suppressor tRNA, is able to selectively incorporate 18 unnatural amino acids into proteins, including trifluoroketone-, alkynyl-, and halogen-substituted amino acids. In an attempt to improve our understanding of this polyspecificity, the X-ray crystal structure of the aaRS-p-cyanophenylalanine complex was determined. A comparison of this structure with those of other mutant aaRSs showed that both binding site size and other more subtle features control substrate polyspecificity.
Figures

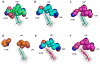
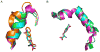
References
-
- Liu CC, Schultz PG. An Expanding Genetic Code. Annual Review of Biochemistry. 2010:79. - PubMed
-
- Wang L, Schultz PG. A general approach for the generation of orthogonal tRNAs. Chem Biol. 2001;8:883–890. - PubMed
-
- Santoro SW, Wang L, Herberich B, King DS, Schultz PG. An efficient system for the evolution of aminoacyl-tRNA synthetase specificity. Nat Biotechnol. 2002;20:1044–1048. - PubMed
-
- Stokes AL, Miyake-Stoner SJ, Peeler JC, Nguyen DP, Hammer RP, Mehl RA. Enhancing the utility of unnatural amino acid synthetases by manipulating broad substrate specificity. Mol Biosyst. 2009;5:1032–1038. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

