Draft genome of the red harvester ant Pogonomyrmex barbatus
- PMID: 21282651
- PMCID: PMC3078412
- DOI: 10.1073/pnas.1007901108
Draft genome of the red harvester ant Pogonomyrmex barbatus
Abstract
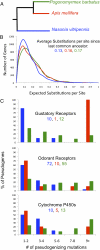
We report the draft genome sequence of the red harvester ant, Pogonomyrmex barbatus. The genome was sequenced using 454 pyrosequencing, and the current assembly and annotation were completed in less than 1 y. Analyses of conserved gene groups (more than 1,200 manually annotated genes to date) suggest a high-quality assembly and annotation comparable to recently sequenced insect genomes using Sanger sequencing. The red harvester ant is a model for studying reproductive division of labor, phenotypic plasticity, and sociogenomics. Although the genome of P. barbatus is similar to other sequenced hymenopterans (Apis mellifera and Nasonia vitripennis) in GC content and compositional organization, and possesses a complete CpG methylation toolkit, its predicted genomic CpG content differs markedly from the other hymenopterans. Gene networks involved in generating key differences between the queen and worker castes (e.g., wings and ovaries) show signatures of increased methylation and suggest that ants and bees may have independently co-opted the same gene regulatory mechanisms for reproductive division of labor. Gene family expansions (e.g., 344 functional odorant receptors) and pseudogene accumulation in chemoreception and P450 genes compared with A. mellifera and N. vitripennis are consistent with major life-history changes during the adaptive radiation of Pogonomyrmex spp., perhaps in parallel with the development of the North American deserts.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
-
The birth of ant genomics.Proc Natl Acad Sci U S A. 2011 Apr 5;108(14):5477-8. doi: 10.1073/pnas.1100765108. Epub 2011 Mar 28. Proc Natl Acad Sci U S A. 2011. PMID: 21444823 Free PMC article. No abstract available.
References
-
- Maynard Smith J, Szathmáry E. The Major Transitions in Evolution. New York: W. H. Freeman/Spektrum; 1995.
-
- Hölldobler B, Wilson EO. The Ants. Cambridge, MA: Belknap Press of Harvard University Press; 1990.
-
- Smith CR, Toth AL, Suarez AV, Robinson GE. Genetic and genomic analyses of the division of labour in insect societies. Nat Rev Genet. 2008;9:735–748. - PubMed
-
- Robinson GE, Grozinger CM, Whitfield CW. Sociogenomics: Social life in molecular terms. Nat Rev Genet. 2005;6:257–270. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

