Comparative genomic analysis of the Streptococcus dysgalactiae species group: gene content, molecular adaptation, and promoter evolution
- PMID: 21282711
- PMCID: PMC3056289
- DOI: 10.1093/gbe/evr006
Comparative genomic analysis of the Streptococcus dysgalactiae species group: gene content, molecular adaptation, and promoter evolution
Abstract
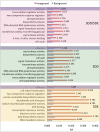
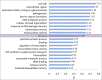
Comparative genomics of closely related bacterial species with different pathogenesis and host preference can provide a means of identifying the specifics of adaptive differences. Streptococcus dysgalactiae (SD) is comprised of two subspecies: S. dysgalactiae subsp. equisimilis is both a human commensal organism and a human pathogen, and S. dysgalactiae subsp. dysgalactiae is strictly an animal pathogen. Here, we present complete genome sequences for both taxa, with analyses involving other species of Streptococcus but focusing on adaptation in the SD species group. We found little evidence for enrichment in biochemical categories of genes carried by each SD strain, however, differences in the virulence gene repertoire were apparent. Some of the differences could be ascribed to prophage and integrative conjugative elements. We identified approximately 9% of the nonrecombinant core genome to be under positive selection, some of which involved known virulence factors in other bacteria. Analyses of proteomes by pooling data across genes, by biochemical category, clade, or branch, provided evidence for increased rates of evolution in several gene categories, as well as external branches of the tree. Promoters were primarily evolving under purifying selection but with certain categories of genes evolving faster. Many of these fast-evolving categories were the same as those associated with rapid evolution in proteins. Overall, these results suggest that adaptation to changing environments and new hosts in the SD species group has involved the acquisition of key virulence genes along with selection of orthologous protein-coding loci and operon promoters.
Figures








References
-
- Abdelsalam M, Chen SC, Yoshida T. Dissemination of streptococcal pyrogenic exotoxin G (spegg) with an IS-like element in fish isolates of Streptococcus dysgalactiae. FEMS Microbiol Lett. 2010;309:105–113. - PubMed
-
- Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. - PubMed
-
- Arakawa K, et al. G-language Genome Analysis Environment: a workbench for nucleotide sequence data mining. Bioinformatics. 2003;19:305–306. - PubMed
-
- Arakawa K, Tomita M. G-language System as a platform for large-scale analysis of high-throughput omics data. J Pesticide Sci. 2006;31:282–288.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

