Bacterial aptamers that selectively bind glutamine
- PMID: 21282981
- PMCID: PMC3127080
- DOI: 10.4161/rna.8.1.13864
Bacterial aptamers that selectively bind glutamine
Abstract
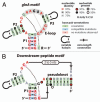
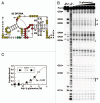
The continued expansion of microbial sequence data has allowed for the detection of an increasing number of conserved RNA motifs by using comparative sequence analysis. Recently, we reported the discovery of two structured non-coding RNA motifs, called glnA and Downstream-peptide, that have similarity in sequence and secondary structure. In this report, we describe data demonstrating that representatives of both RNA motifs selectively bind the amino acid L-glutamine. These glutamine aptamers are found exclusively in cyanobacteria and marine metagenomic sequences, wherein several glnA RNA representatives reside upstream of genes involved in nitrogen metabolism. These motifs have genomic distributions that are consistent with a gene regulation function, suggesting they are components of glutamine-responsive riboswitches. Thus, our findings implicate glutamine as a regulator of cyanobacterial nitrogen metabolism pathways. Furthermore, our findings expand the collection of natural aptamer classes that bind amino acids to include glycine, lysine and glutamine.
Figures


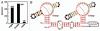
References
-
- Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, et al. A trans-acting riboswitch controls expression of the virulence regulator PrfA in listeria monocytogenes. Cell. 2009;139:770–779. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
