Fine mapping of the NRG1 Hirschsprung's disease locus
- PMID: 21283760
- PMCID: PMC3024406
- DOI: 10.1371/journal.pone.0016181
Fine mapping of the NRG1 Hirschsprung's disease locus
Abstract
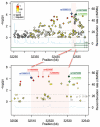
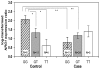
The primary pathology of Hirschsprung's disease (HSCR, colon aganglionosis) is the absence of ganglia in variable lengths of the hindgut, resulting in functional obstruction. HSCR is attributed to a failure of migration of the enteric ganglion precursors along the developing gut. RET is a key regulator of the development of the enteric nervous system (ENS) and the major HSCR-causing gene. Yet the reduced penetrance of RET DNA HSCR-associated variants together with the phenotypic variability suggest the involvement of additional genes in the disease. Through a genome-wide association study, we uncovered a ∼350 kb HSCR-associated region encompassing part of the neuregulin-1 gene (NRG1). To identify the causal NRG1 variants contributing to HSCR, we genotyped 243 SNPs variants on 343 ethnic Chinese HSCR patients and 359 controls. Genotype analysis coupled with imputation narrowed down the HSCR-associated region to 21 kb, with four of the most associated SNPs (rs10088313, rs10094655, rs4624987, and rs3884552) mapping to the NRG1 promoter. We investigated whether there was correlation between the genotype at the rs10088313 locus and the amount of NRG1 expressed in human gut tissues (40 patients and 21 controls) and found differences in expression as a function of genotype. We also found significant differences in NRG1 expression levels between diseased and control individuals bearing the same rs10088313 risk genotype. This indicates that the effects of NRG1 common variants are likely to depend on other alleles or epigenetic factors present in the patients and would account for the variability in the genetic predisposition to HSCR.
Conflict of interest statement
Figures
References
-
- Amiel J, Sproat-Emison E, Garcia-Barcelo M, Lantieri F, Burzynski G, et al. Hirschsprung disease, associated syndromes and genetics: a review. J MedGenet. 2008;45:1–14. - PubMed
-
- Torfs CP. An epidemiological study of Hirschsprung disease in a multiracial California population; 1998; Evian, France.
-
- Edery P, Lyonnet S, Mulligan LM, Pelet A, Dow E, et al. Mutations of the RET proto-oncogene in Hirschsprung's disease. Nature. 1994;367:378–380. - PubMed
-
- Romeo G, Ronchetto P, Luo Y, Barone V, Seri M, et al. Point mutations affecting the tyrosine kinase domain of the RET proto-oncogene in Hirschsprung's disease. Nature. 1994;367:377–378. - PubMed
-
- Hofstra RM, Wu Y, Stulp RP, Elfferich P, Osinga J, et al. RET and GDNF gene scanning in Hirschsprung patients using two dual denaturing gel systems. Hum Mutat. 2000;15:418–429. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

