Characterization of the deleted in autism 1 protein family: implications for studying cognitive disorders
- PMID: 21283809
- PMCID: PMC3023760
- DOI: 10.1371/journal.pone.0014547
Characterization of the deleted in autism 1 protein family: implications for studying cognitive disorders
Abstract
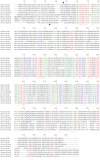
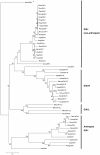
Autism spectrum disorders (ASDs) are a group of commonly occurring, highly-heritable developmental disabilities. Human genes c3orf58 or Deleted In Autism-1 (DIA1) and cXorf36 or Deleted in Autism-1 Related (DIA1R) are implicated in ASD and mental retardation. Both gene products encode signal peptides for targeting to the secretory pathway. As evolutionary medicine has emerged as a key tool for understanding increasing numbers of human diseases, we have used an evolutionary approach to study DIA1 and DIA1R. We found DIA1 conserved from cnidarians to humans, indicating DIA1 evolution coincided with the development of the first primitive synapses. Nematodes lack a DIA1 homologue, indicating Caenorhabditis elegans is not suitable for studying all aspects of ASD etiology, while zebrafish encode two DIA1 paralogues. By contrast to DIA1, DIA1R was found exclusively in vertebrates, with an origin coinciding with the whole-genome duplication events occurring early in the vertebrate lineage, and the evolution of the more complex vertebrate nervous system. Strikingly, DIA1R was present in schooling fish but absent in fish that have adopted a more solitary lifestyle. An additional DIA1-related gene we named DIA1-Like (DIA1L), lacks a signal peptide and is restricted to the genomes of the echinoderm Strongylocentrotus purpuratus and cephalochordate Branchiostoma floridae. Evidence for remarkable DIA1L gene expansion was found in B. floridae. Amino acid alignments of DIA1 family gene products revealed a potential Golgi-retention motif and a number of conserved motifs with unknown function. Furthermore, a glycine and three cysteine residues were absolutely conserved in all DIA1-family proteins, indicating a critical role in protein structure and/or function. We have therefore identified a new metazoan protein family, the DIA1-family, and understanding the biological roles of DIA1-family members will have implications for our understanding of autism and mental retardation.
Conflict of interest statement
Figures




References
-
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77. - PubMed
-
- Folstein SE, Rosen-Sheidley B. Genetics of autism: complex aetiology for a heterogeneous disorder. Nat Rev Genet. 2001;2:943–955. - PubMed
-
- Veenstra-VanderWeele J, Cook EH., Jr Molecular genetics of autism spectrum disorder. Mol. Psychiatry. 2004;9:819–832. - PubMed
-
- Rutter M. Genetic studies of autism: from the 1970s into the millennium. J Abnorm Child Psychol. 2000;28:3–14. - PubMed
-
- Ronald A, Happé F, Bolton P, Butcher LM, Price TS, et al. Genetic heterogeneity between the three components of the autism spectrum: a twin study. J Am Acad Child Adolesc Psychiatry. 2006;45:691–699. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

