Stabilization by fusion to the C-terminus of hyperthermophile Sulfolobus tokodaii RNase HI: a possibility of protein stabilization tag
- PMID: 21283826
- PMCID: PMC3023800
- DOI: 10.1371/journal.pone.0016226
Stabilization by fusion to the C-terminus of hyperthermophile Sulfolobus tokodaii RNase HI: a possibility of protein stabilization tag
Abstract
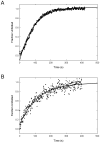
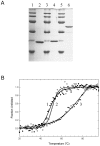
RNase HI from the hyperthermophile Sulfolobus tokodaii (Sto-RNase HI) is stabilized by its C-terminal residues. In this work, the stabilization effect of the Sto-RNase HI C-terminal residues was investigated in detail by thermodynamic measurements of the stability of variants lacking the disulfide bond (C58/145A), or the six C-terminal residues (ΔC6) and by structural analysis of ΔC6. The results showed that the C-terminal does not affect overall structure and stabilization is caused by local interactions of the C-terminal, suggesting that the C-terminal residues could be used as a "stabilization tag." The Sto-RNase HI C-terminal residues (-IGCIILT) were introduced as a tag on three proteins. Each chimeric protein was more stable than its wild-type protein. These results suggested the possibility of a simple stabilization technique using a stabilization tag such as Sto-RNase HI C-terminal residues.
Conflict of interest statement
Figures




References
-
- Kimura S, Nakamura H, Hashimoto T, Oobatake M, Kanaya S. Stabilization of Escherichia coli ribonuclease HI by strategic replacement of amino acid residues with those from the thermophilic counterpart. J Biol Chem. 1992;267:21535–21542. - PubMed
-
- Watanabe K, Ohkuri T, Yokobori S, Yamagishi A. Designing thermostable proteins: ancestral mutants of 3-isopropylmalate dehydrogenase designed by using a phylogenetic tree. J Mol Biol. 2006;355:664–674. - PubMed
-
- Miyazaki K, Wintrode PL, Grayling RA, Rubingh DN, Arnold FH. Directed evolution study of temperature adaptation in a psychrophilic enzyme. J Mol Biol. 2000;297:1015–1026. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

