Genomewide analyses define different modes of transcriptional regulation by peroxisome proliferator-activated receptor-β/δ (PPARβ/δ)
- PMID: 21283829
- PMCID: PMC3023804
- DOI: 10.1371/journal.pone.0016344
Genomewide analyses define different modes of transcriptional regulation by peroxisome proliferator-activated receptor-β/δ (PPARβ/δ)
Abstract
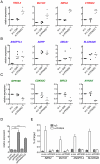
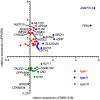
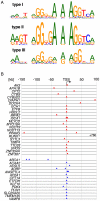
Peroxisome proliferator-activated receptors (PPARs) are nuclear receptors with essential functions in lipid, glucose and energy homeostasis, cell differentiation, inflammation and metabolic disorders, and represent important drug targets. PPARs heterodimerize with retinoid X receptors (RXRs) and can form transcriptional activator or repressor complexes at specific DNA elements (PPREs). It is believed that the decision between repression and activation is generally governed by a ligand-mediated switch. We have performed genomewide analyses of agonist-treated and PPARβ/δ-depleted human myofibroblasts to test this hypothesis and to identify global principles of PPARβ/δ-mediated gene regulation. Chromatin immunoprecipitation sequencing (ChIP-Seq) of PPARβ/δ, H3K4me3 and RNA polymerase II enrichment sites combined with transcriptional profiling enabled the definition of 112 bona fide PPARβ/δ target genes showing either of three distinct types of transcriptional response: (I) ligand-independent repression by PPARβ/δ; (II) ligand-induced activation and/or derepression by PPARβ/δ; and (III) ligand-independent activation by PPARβ/δ. These data identify PPRE-mediated repression as a major mechanism of transcriptional regulation by PPARβ/δ, but, unexpectedly, also show that only a subset of repressed genes are activated by a ligand-mediated switch. Our results also suggest that the type of transcriptional response by a given target gene is connected to the structure of its associated PPRE(s) and the biological function of its encoded protein. These observations have important implications for understanding the regulatory PPAR network and PPARβ/δ ligand-based drugs.
Conflict of interest statement
Figures


References
-
- Desvergne B, Michalik L, Wahli W. Transcriptional regulation of metabolism. Physiol Rev. 2006;86:465–514. - PubMed
-
- Kilgore KS, Billin AN. PPARbeta/delta ligands as modulators of the inflammatory response. Curr Opin Investig Drugs. 2008;9:463–469. - PubMed
-
- Grimaldi PA. Metabolic and nonmetabolic regulatory functions of peroxisome proliferator-activated receptor beta. Curr Opin Lipidol. 2010;21:186–191. - PubMed
-
- Peraza MA, Burdick AD, Marin HE, Gonzalez FJ, Peters JM. The toxicology of ligands for peroxisome proliferator-activated receptors (PPAR). Toxicol Sci. 2006;90:269–295. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

