A microfabricated electrical differential counter for the selective enumeration of CD4+ T lymphocytes
- PMID: 21283908
- PMCID: PMC3141315
- DOI: 10.1039/c0lc00556h
A microfabricated electrical differential counter for the selective enumeration of CD4+ T lymphocytes
Abstract
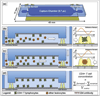
We have developed a microfabricated biochip that enumerates CD4+ T lymphocytes from leukocyte populations obtained from human blood samples using electrical impedance sensing and immunoaffinity chromatography. T cell counts are found by obtaining the difference between the number of leukocytes before and after depleting CD4+ T cells with immobilized antibodies in a capture chamber. This differential counting technique has been validated to analyze physiological concentrations of leukocytes with an accuracy of ∼9 cells per µL by passivating the capture chamber with bovine serum albumin. In addition, the counter provided T cell counts which correlated closely with an optical control (R(2) = 0.997) for CD4 cell concentrations ranging from approximately 100 to 700 cells per µL. We believe that this approach can be a promising method for bringing quantitative HIV/AIDS diagnostics to resource-poor regions in the world.
Figures






Similar articles
-
Coincidence detection of heterogeneous cell populations from whole blood with coplanar electrodes in a microfluidic impedance cytometer.Lab Chip. 2014 Nov 21;14(22):4370-81. doi: 10.1039/c4lc00879k. Lab Chip. 2014. PMID: 25231594
-
A multicenter evaluation of the PanLeucogating method and the use of generic monoclonal antibody reagents for CD4 enumeration in HIV-infected patients in Thailand.Cytometry B Clin Cytom. 2005 May;65(1):29-36. doi: 10.1002/cyto.b.20052. Cytometry B Clin Cytom. 2005. PMID: 15800883 Clinical Trial.
-
Cell detection and counting through cell lysate impedance spectroscopy in microfluidic devices.Lab Chip. 2007 Jun;7(6):746-55. doi: 10.1039/b705082h. Epub 2007 May 11. Lab Chip. 2007. PMID: 17538717 Free PMC article.
-
CD4 counting technologies for HIV therapy monitoring in resource-poor settings--state-of-the-art and emerging microtechnologies.Lab Chip. 2013 Jul 21;13(14):2731-48. doi: 10.1039/c3lc50213a. Lab Chip. 2013. PMID: 23670110 Review.
-
Absolute and percent CD4+ T-cell enumeration by flow cytometry using capillary blood.J Immunol Methods. 2011 Sep 30;372(1-2):1-6. doi: 10.1016/j.jim.2011.07.008. Epub 2011 Jul 20. J Immunol Methods. 2011. PMID: 21787779 Review.
Cited by
-
Thin flexible lab-on-a-film for impedimetric sensing in biomedical applications.Sci Rep. 2022 Jan 20;12(1):1066. doi: 10.1038/s41598-022-04917-5. Sci Rep. 2022. PMID: 35058505 Free PMC article.
-
On a chip.IEEE Pulse. 2011 Nov;2(6):19-27. doi: 10.1109/MPUL.2011.942762. IEEE Pulse. 2011. PMID: 22147065 Free PMC article.
-
Optofluidic fluorescent imaging cytometry on a cell phone.Anal Chem. 2011 Sep 1;83(17):6641-7. doi: 10.1021/ac201587a. Epub 2011 Aug 2. Anal Chem. 2011. PMID: 21774454 Free PMC article.
-
Microfluidic diagnostic tool for the developing world: contactless impedance flow cytometry.Lab Chip. 2012 Nov 7;12(21):4499-507. doi: 10.1039/c2lc40759k. Lab Chip. 2012. PMID: 22971813 Free PMC article.
-
CMOS cell sensors for point-of-care diagnostics.Sensors (Basel). 2012;12(8):10042-66. doi: 10.3390/s120810042. Epub 2012 Jul 25. Sensors (Basel). 2012. PMID: 23112587 Free PMC article. Review.
References
-
- UNAIDS and WHO. [accessed 7 October, 2010];AIDS Epidemic Update. 2009 http://data.unaids.org/pub/Report/2009/jc1700_epi_update_2009_en.pdf.
-
- Kitahata M, Gange S, Abraham A, Merriman B, Saag M, Justice A, Hogg R, Deeks S, Eron J, Brooks J, Rourke S, Gill M, Bosch R, Martin J, Klein M, Jacobson L, Rodriguez B, Sterling T, Kirk G, Napravnik S, Rachlis A, Calzavara L, Horgerg M, Silverberg M, Gebo K, Goedert J, Benson C, Collier A, Van Rompaey S, Crane H, McKaig R, Lau B, Freeman A, Moore R. N. Engl. J. Med. 2009;360:1815. - PMC - PubMed
-
- WHO, UNICEF, UNAIDS. [accessed 7 October, 2010];Towards Universal Access: Scaling Up Priority HIV/AIDS Interventions in the Health Sector. http://www.who.int/entity/hiv/pub/tuapr_2009_en.pdf.
-
- Phillips A, Lee C, Elford J, Janossy G, Timms A, Bofill M, Kernoff P. Lancet. 1991;337:389. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

