Phenotype harmonization and cross-study collaboration in GWAS consortia: the GENEVA experience
- PMID: 21284036
- PMCID: PMC3055921
- DOI: 10.1002/gepi.20564
Phenotype harmonization and cross-study collaboration in GWAS consortia: the GENEVA experience
Abstract
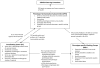
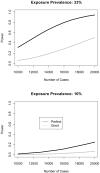
Genome-wide association study (GWAS) consortia and collaborations formed to detect genetic loci for common phenotypes or investigate gene-environment (G*E) interactions are increasingly common. While these consortia effectively increase sample size, phenotype heterogeneity across studies represents a major obstacle that limits successful identification of these associations. Investigators are faced with the challenge of how to harmonize previously collected phenotype data obtained using different data collection instruments which cover topics in varying degrees of detail and over diverse time frames. This process has not been described in detail. We describe here some of the strategies and pitfalls associated with combining phenotype data from varying studies. Using the Gene Environment Association Studies (GENEVA) multi-site GWAS consortium as an example, this paper provides an illustration to guide GWAS consortia through the process of phenotype harmonization and describes key issues that arise when sharing data across disparate studies. GENEVA is unusual in the diversity of disease endpoints and so the issues it faces as its participating studies share data will be informative for many collaborations. Phenotype harmonization requires identifying common phenotypes, determining the feasibility of cross-study analysis for each, preparing common definitions, and applying appropriate algorithms. Other issues to be considered include genotyping timeframes, coordination of parallel efforts by other collaborative groups, analytic approaches, and imputation of genotype data. GENEVA's harmonization efforts and policy of promoting data sharing and collaboration, not only within GENEVA but also with outside collaborations, can provide important guidance to ongoing and new consortia.
© 2011 Wiley-Liss, Inc.
Figures
References
-
- Cornelis MC, Agrawal A, Cole JW, Hansel NN, Barnes KC, Beaty TH, Bennett SN, Bierut LJ, Boerwinkle E, Doheny KF, Feenstra B, Feingold E, Fornage M, Haiman CA, Harris EL, Hayes MG, Heit JA, Hu FB, Kang JH, Laurie CC, Ling H, Manolio TA, Marazita ML, Mathias RA, Mirel DB, Paschall J, Pasquale LR, Pugh EW, Rice JP, Udren J, van Dam RM, Wang X, Wiggs JL, Williams K, Yu K, for the GENEVA Consortium The Gene, Environment Association Studies Consortium (GENEVA): Maximizing the knowledge obtained from GWAS by collaboration across studies of multiple conditions. Genet Epidemiol. 2010;34:364–372. - PMC - PubMed
-
- Garcia-Closas M, Lubin JH. Power and sample size calculations in case-control studies of gene-environment interactions: Comments on different approaches. Am J Epidemiol. 1999;149:689–692. - PubMed
Publication types
MeSH terms
Grants and funding
- R01HL59367/HL/NHLBI NIH HHS/United States
- Z01 CP010200/ImNIH/Intramural NIH HHS/United States
- R01NS45012/NS/NINDS NIH HHS/United States
- N01 HC055015/HL/NHLBI NIH HHS/United States
- U01 HG004415/HG/NHGRI NIH HHS/United States
- HHSN268200782096C/HG/NHGRI NIH HHS/United States
- R01 DA013423/DA/NIDA NIH HHS/United States
- R01 CA063464/CA/NCI NIH HHS/United States
- U01 HG004735/HG/NHGRI NIH HHS/United States
- U01 CA136792/CA/NCI NIH HHS/United States
- U01HG004446/HG/NHGRI NIH HHS/United States
- UL1RR025005/RR/NCRR NIH HHS/United States
- CA54281/CA/NCI NIH HHS/United States
- N01HC-55021/HC/NHLBI NIH HHS/United States
- P01CA089392/CA/NCI NIH HHS/United States
- U01HG004438/HG/NHGRI NIH HHS/United States
- U01HG004726/HG/NHGRI NIH HHS/United States
- N01 HC055016/HL/NHLBI NIH HHS/United States
- R01 NS045012/NS/NINDS NIH HHS/United States
- R01 DK058845/DK/NIDDK NIH HHS/United States
- U01 HG004728/HG/NHGRI NIH HHS/United States
- U01 HG004438/HG/NHGRI NIH HHS/United States
- U10AA008401/AA/NIAAA NIH HHS/United States
- U01 HG004446/HG/NHGRI NIH HHS/United States
- N01 HC055019/HL/NHLBI NIH HHS/United States
- UL1 RR025005/RR/NCRR NIH HHS/United States
- U01HG004738/HG/NHGRI NIH HHS/United States
- U01HG004415/HG/NHGRI NIH HHS/United States
- R01EY015473/EY/NEI NIH HHS/United States
- U01 DE018993/DE/NIDCR NIH HHS/United States
- R01 HL059367/HL/NHLBI NIH HHS/United States
- U01HG004735/HG/NHGRI NIH HHS/United States
- N01 HC055021/HL/NHLBI NIH HHS/United States
- P01 CA089392/CA/NCI NIH HHS/United States
- CAPMC/ CIHR/Canada
- R01 DE014899/DE/NIDCR NIH HHS/United States
- R01 HL086694/HL/NHLBI NIH HHS/United States
- U01 HG004402/HG/NHGRI NIH HHS/United States
- U01 HG004424/HG/NHGRI NIH HHS/United States
- U01HG004728/HG/NHGRI NIH HHS/United States
- U01HG004399/HG/NHGRI NIH HHS/United States
- U01HG004402/HG/NHGRI NIH HHS/United States
- U01HG004422/HG/NHGRI NIH HHS/United States
- CA63464/CA/NCI NIH HHS/United States
- N01 HC055020/HL/NHLBI NIH HHS/United States
- R01DA013423/DA/NIDA NIH HHS/United States
- U01 HG004729/HG/NHGRI NIH HHS/United States
- P30 DK072488/DK/NIDDK NIH HHS/United States
- U01 HG004422/HG/NHGRI NIH HHS/United States
- U01HG04424/HG/NHGRI NIH HHS/United States
- R01HL087641/HL/NHLBI NIH HHS/United States
- U01 NS069208/NS/NINDS NIH HHS/United States
- U01DE018993/DE/NIDCR NIH HHS/United States
- U01HG004423/HG/NHGRI NIH HHS/United States
- R01 EY015473/EY/NEI NIH HHS/United States
- R01 CA054281/CA/NCI NIH HHS/United States
- U01DE018903/DE/NIDCR NIH HHS/United States
- U01 CA063464/CA/NCI NIH HHS/United States
- N01 HC055018/HL/NHLBI NIH HHS/United States
- U01HG004436/HG/NHGRI NIH HHS/United States
- U01HG004729/HG/NHGRI NIH HHS/United States
- U01 DE018903/DE/NIDCR NIH HHS/United States
- U01 HG004436/HG/NHGRI NIH HHS/United States
- N01 HC055022/HL/NHLBI NIH HHS/United States
- U01 HG004738/HG/NHGRI NIH HHS/United States
- U10 AA008401/AA/NIAAA NIH HHS/United States
- R01 EY015872/EY/NEI NIH HHS/United States
- U01 HG004423/HG/NHGRI NIH HHS/United States
- N01HC-55020/HC/NHLBI NIH HHS/United States
- R01 HL087641/HL/NHLBI NIH HHS/United States
- R37 CA054281/CA/NCI NIH HHS/United States
- U01 HG004399/HG/NHGRI NIH HHS/United States
- CA136792/CA/NCI NIH HHS/United States
- R01EY015872/EY/NEI NIH HHS/United States
- R01HL086694/HL/NHLBI NIH HHS/United States
- U01 HG004726/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

