The role of codon selection in regulation of translation efficiency deduced from synthetic libraries
- PMID: 21284851
- PMCID: PMC3188794
- DOI: 10.1186/gb-2011-12-2-r12
The role of codon selection in regulation of translation efficiency deduced from synthetic libraries
Abstract
Background: Translation efficiency is affected by a diversity of parameters, including secondary structure of the transcript and its codon usage. Here we examine the effects of codon usage on translation efficiency by re-analysis of previously constructed synthetic expression libraries in Escherichia coli.
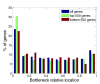
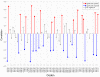
Results: We define the region in a gene that takes the longest time to translate as the bottleneck. We found that localization of the bottleneck at the beginning of a transcript promoted a high level of expression, especially if the computed dwell time of the ribosome within this region was sufficiently long. The location and translation time of the bottleneck were not correlated with the cost of expression, approximated by the fitness of the host cell, yet utilization of specific codons was. Particularly, enhanced usage of the codons UCA and CAU was correlated with increased cost of production, potentially due to sequestration of their corresponding rare tRNAs.
Conclusions: The distribution of codons along the genes appears to affect translation efficiency, consistent with analysis of natural genes. This study demonstrates how synthetic biology complements bioinformatics by providing a set-up for well controlled experiments in biology.
Figures
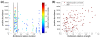


References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

