Neutral evolution of robustness in Drosophila microRNA precursors
- PMID: 21285032
- PMCID: PMC3167684
- DOI: 10.1093/molbev/msr029
Neutral evolution of robustness in Drosophila microRNA precursors
Abstract
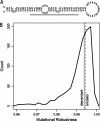
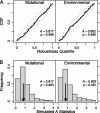
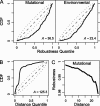
Mutational robustness describes the extent to which a phenotype remains unchanged in the face of mutations. Theory predicts that the strength of direct selection for mutational robustness is at most the magnitude of the rate of deleterious mutation. As far as nucleic acid sequences are concerned, only long sequences in organisms with high deleterious mutation rates and large population sizes are expected to evolve mutational robustness. Surprisingly, recent studies have concluded that molecules that meet none of these conditions--the microRNA precursors (pre-miRNAs) of multicellular eukaryotes--show signs of selection for mutational and/or environmental robustness. To resolve the apparent disagreement between theory and these studies, we have reconstructed the evolutionary history of Drosophila pre-miRNAs and compared the robustness of each sequence to that of its reconstructed ancestor. In addition, we "replayed the tape" of pre-miRNA evolution via simulation under different evolutionary assumptions and compared these alternative histories with the actual one. We found that Drosophila pre-miRNAs have evolved under strong purifying selection against changes in secondary structure. Contrary to earlier claims, there is no evidence that these RNAs have been shaped by either direct or congruent selection for any kind of robustness. Instead, the high robustness of Drosophila pre-miRNAs appears to be mostly intrinsic and likely a consequence of selection for functional structures.
Figures


References
-
- Ancel LW, Fontana W. Plasticity, evolvability, and modularity in RNA. J Exp Zool (Mol Dev Evol) 2000;288:242–283. - PubMed
-
- Anderson TW, Darling DA. Asymptotic theory of certain “goodness of fit” criteria based on stochastic processes. Ann Math Statist. 1952;23:193–212.
-
- Azevedo RBR, Lohaus R, Srinivasan S, Dang KK, Burch CL. Sexual reproduction selects for robustness and negative epistasis in artificial gene networks. Nature. 2006;440:87–90. - PubMed
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Blanchette M, Diallo AB, Green ED, Miller W, Haussler D. Computational reconstruction of ancestral DNA sequences. Methods Mol Biol. 2008;422:171–184. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

