The R2R3 MYB transcription factor DUO1 activates a male germline-specific regulon essential for sperm cell differentiation in Arabidopsis
- PMID: 21285328
- PMCID: PMC3077786
- DOI: 10.1105/tpc.110.081059
The R2R3 MYB transcription factor DUO1 activates a male germline-specific regulon essential for sperm cell differentiation in Arabidopsis
Abstract
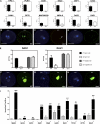
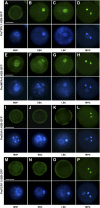
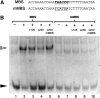
The male germline in flowering plants arises through asymmetric division of a haploid microspore. The resulting germ cell undergoes mitotic division and specialization to produce the two sperm cells required for double fertilization. The male germline-specific R2R3 MYB transcription factor DUO1 POLLEN1 (DUO1) plays an essential role in sperm cell specification by activating a germline-specific differentiation program. Here, we show that ectopic expression of DUO1 upregulates a significant number (~63) of germline-specific or enriched genes, including those required for fertilization. We validated 14 previously unknown DUO1 target genes by demonstrating DUO1-dependent promoter activity in the male germline. DUO1 is shown to directly regulate its target promoters through binding to canonical MYB sites, suggesting that the DUO1 target genes validated thus far are likely to be direct targets. This work advances knowledge of the DUO1 regulon that encompasses genes with a range of cellular functions, including transcription, protein fate, signaling, and transport. Thus, the DUO1 regulon has a major role in shaping the germline transcriptome and functions to commit progenitor germ cells to sperm cell differentiation.
Figures



References
-
- Baek K.H. (1999). The first oncogene in Drosophila melanogaster. Mutat. Res. 436: 131–136 - PubMed
-
- Bell S.D., Botting C.H., Wardleworth B.N., Jackson S.P., White M.F. (2002). The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science 296: 148–151 - PubMed
-
- Berger F., Hamamura Y., Ingouff M., Higashiyama T. (2008). Double fertilization - caught in the act. Trends Plant Sci. 13: 437–443 - PubMed
-
- Bilder D. (2001). Cell polarity: Squaring the circle. Curr. Biol. 11: R132–R135 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

