The nature of protein domain evolution: shaping the interaction network
- PMID: 21286315
- PMCID: PMC2945003
- DOI: 10.2174/138920210791616725
The nature of protein domain evolution: shaping the interaction network
Abstract
The proteomes that make up the collection of proteins in contemporary organisms evolved through recombination and duplication of a limited set of domains. These protein domains are essentially the main components of globular proteins and are the most principal level at which protein function and protein interactions can be understood. An important aspect of domain evolution is their atomic structure and biochemical function, which are both specified by the information in the amino acid sequence. Changes in this information may bring about new folds, functions and protein architectures. With the present and still increasing wealth of sequences and annotation data brought about by genomics, new evolutionary relationships are constantly being revealed, unknown structures modeled and phylogenies inferred. Such investigations not only help predict the function of newly discovered proteins, but also assist in mapping unforeseen pathways of evolution and reveal crucial, co-evolving inter- and intra-molecular interactions. In turn this will help us describe how protein domains shaped cellular interaction networks and the dynamics with which they are regulated in the cell. Additionally, these studies can be used for the design of new and optimized protein domains for therapy. In this review, we aim to describe the basic concepts of protein domain evolution and illustrate recent developments in molecular evolution that have provided valuable new insights in the field of comparative genomics and protein interaction networks.
Keywords: PDZ domain; Protein domain; interactome.; molecular evolution; superfamily; systems biology.
Figures

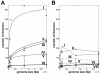
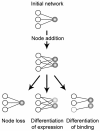
References
-
- Ladunga I. Phylogenetic continuum indicates "galaxies" in the protein universe: perliminary results on the natural group structures of proteins. J. Mol. Evol. 1992;34:358–375. - PubMed
-
- Bailey K, Sanger F. The chemistry of amino acids and proteins. Annu. Rev. Biochem. 1951;20:103–130. - PubMed
-
- Sanger F. Some peptides from insulin. Nature. 1948;162:491. - PubMed
-
- Adams JM, Jeppesen PG, Sanger F, Barrell BG. Nucleotide sequence from the coat protein cistron of R17 bacteriophage RNA. Nature. 1969;223:1009–1014. - PubMed
LinkOut - more resources
Full Text Sources
