Structural regulation of cullin-RING ubiquitin ligase complexes
- PMID: 21288713
- PMCID: PMC3151539
- DOI: 10.1016/j.sbi.2011.01.003
Structural regulation of cullin-RING ubiquitin ligase complexes
Abstract
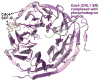
Cullin-RING ligases (CRLs) compose the largest class of E3 ubiquitin ligases. CRLs are modular, multisubunit enzymes, comprising interchangeable substrate receptors dedicated to particular Cullin-RING catalytic cores. Recent structural studies have revealed numerous ways in which CRL E3 ligase activities are controlled, including multimodal E3 ligase activation by covalent attachment of the ubiquitin-like protein NEDD8, inhibition of CRL assembly/activity by CAND1, and several mechanisms of regulated substrate recruitment. These features highlight the potential for CRL activities to be tuned in responses to diverse cellular cues, and for modulating CRL functions through small-molecule agonists or antagonists. As the second installment of a two-review series, this article focuses on recent structural studies advancing our knowledge of how CRL activities are regulated.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors declare no competing interest.
Figures

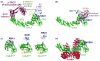

References
-
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. - PubMed
-
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. - PubMed
-
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

