Yeast cells can access distinct quiescent states
- PMID: 21289062
- PMCID: PMC3042157
- DOI: 10.1101/gad.2011311
Yeast cells can access distinct quiescent states
Abstract
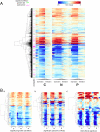
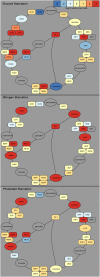
We conducted a phenotypic, transcriptional, metabolic, and genetic analysis of quiescence in yeast induced by starvation of prototrophic cells for one of three essential nutrients (glucose, nitrogen, or phosphate) and compared those results with those obtained with cells growing slowly due to nutrient limitation. These studies address two related questions: (1) Is quiescence a state distinct from any attained during mitotic growth, and (2) does the nature of quiescence differ depending on the means by which it is induced? We found that either limitation or starvation for any of the three nutrients elicits all of the physiological properties associated with quiescence, such as enhanced cell wall integrity and resistance to heat shock and oxidative stress. Moreover, the starvations result in a common transcriptional program, which is in large part a direct extrapolation of the changes that occur during slow growth. In contrast, the metabolic changes that occur upon starvation and the genetic requirements for surviving starvation differ significantly depending on the nutrient for which the cell is starved. The genes needed by cells to survive starvation do not overlap the genes that are induced upon starvation. We conclude that cells do not access a unique and discrete G(0) state, but rather are programmed, when nutrients are scarce, to prepare for a range of possible future stressors. Moreover, these survival strategies are not unique to quiescence, but are engaged by the cell in proportion to nutrient scarcity.
Figures



References
-
- Benjamini Y, Yekutieli D 2001. The control of the false discovery rate in multiple testing under dependency. Ann Stat 29: 1165–1188
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
