Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor
- PMID: 21289069
- PMCID: PMC3034898
- DOI: 10.1101/gad.2001911
Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor
Abstract
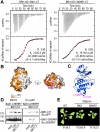
Receptor tyrosine kinases control many critical processes in metazoans, but these enzymes appear to be absent in plants. Recently, two Arabidopsis receptor kinases--BRASSINOSTEROID INSENSITIVE 1 (BRI1) and BRI1-ASSOCIATED KINASE1 (BAK1), the receptor and coreceptor for brassinosteroids--were shown to autophosphorylate on tyrosines. However, the cellular roles for tyrosine phosphorylation in plants remain poorly understood. Here, we report that the BRI1 KINASE INHIBITOR 1 (BKI1) is tyrosine phosphorylated in response to brassinosteroid perception. Phosphorylation occurs within a reiterated [KR][KR] membrane targeting motif, releasing BKI1 into the cytosol and enabling formation of an active signaling complex. Our work reveals that tyrosine phosphorylation is a conserved mechanism controlling protein localization in all higher organisms.
Figures



References
-
- Bayliss R, Sardon T, Vernos I, Conti E 2003. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol Cell 12: 851–862 - PubMed
-
- Belkhadir Y, Chory J 2006. Brassinosteroid signaling: A paradigm for steroid hormone signaling from the cell surface. Science 314: 1410–1411 - PubMed
-
- Cock JM, Vanoosthuyse V, Gaude T 2002. Receptor kinase signalling in plants and animals: Distinct molecular systems with mechanistic similarities. Curr Opin Cell Biol 14: 230–236 - PubMed
-
- Diella F, Haslam N, Chica C, Budd A, Michael S, Brown NP, Trave G, Gibson TJ 2008. Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front Biosci 13: 6580–6603 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
